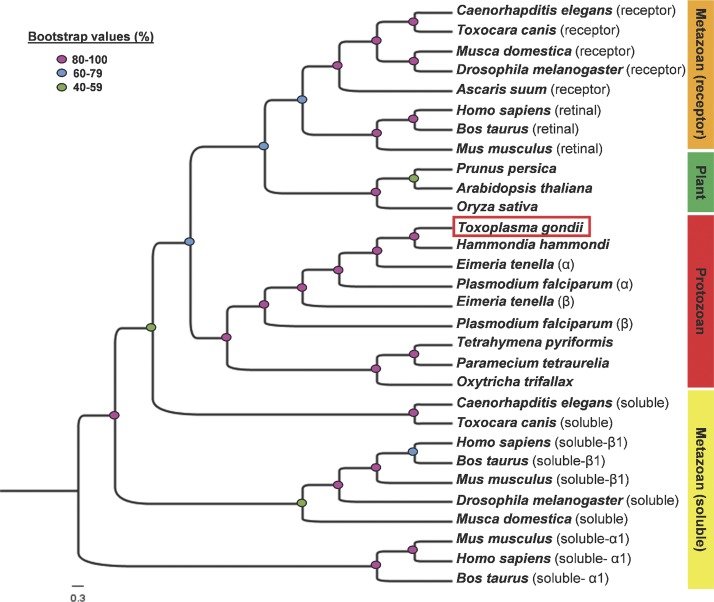
Figure S1. Phylogenetic analysis reveals a protozoan-specific clading of TgATPaseP-GC.
The tree shows the evolutionary relationship of TgATPaseP-GC from T. gondii with orthologs from the listed organisms signifying various domains of life. The image represents a single most parsimonious cladogram generated by maximum likelihood method. Sequence alignment and construction of the tree were performed by CLC Genomics Workbench v12.0, followed by visualization using Figtree v1.4.3. The colored dots on branching nodes show the bootstrap values. Organism abbreviations and accession numbers: Arabidopsis thaliana, AAM51559.1; Ascaris suum (receptor type), PRJNA80881; Bos taurus (retinal type), NP_776973.2; B. taurus (soluble α1-subunit), P19687.1; B. taurus (soluble β1-subunit), P16068.1; Caenorhabditis elegans (receptor type), NP_494995.2; C. elegans (soluble), NP_510557.3; Drosophila melanogaster (receptor type), AAA85858.1; D. melanogaster (soluble β-subunit), NP_524603.2; E. tenella (soluble α), XP_013229212.1; E. tenella (particulate β), XP_013235760.1; Hammondia hammondi, HHA_254370; Homo sapiens (retinal type), NP_000171.1; H. sapiens (soluble α1-subunit), NP_001124157.1; H. sapiens (soluble β1-subunit), NP_001278880.1; Musca domestica (receptor type), XP_005177218.1; M. domestica (soluble), XP_019895151.1; Mus musculus (retinal type), NP_001007577.1; M. musculus (soluble α1-subunit), AAG17446.1; M. musculus (soluble β1-subunit), AAG17447.1; Oryza sativa, ABD18448.1; Oxytricha trifallax, EJY85073.1; Paramecium tetraurelia, XP_001346995.1; P. falciparum (soluble α), AJ245435.1; P. falciparum (particulate β), AJ249165.1; Prunus persica, AGN29346.1; Tetrahymena pyriformis, AJ238858.1; T. gondii, EPR59074.1; Toxocara canis (receptor-type), KHN81453.1; and T. canis (soluble), KHN85312.1.