Abstract
Since publication of the crystal structure of protein kinase (PK)A three decades ago, a structural portrait of the conserved kinase core has been drawn. The next challenge is to elucidate structures of full-length kinases and to address the intrinsically disordered regions (IDRs) that typically flank the core as well as the small linear motifs (SLiMs) that are embedded within the IDRs. It is increasingly apparent that unstructured regions integrate the kinase catalytic chassis into multienzyme-based regulatory networks. The extracellular signal-regulated kinase-ribosomal S6 PK-phosphoinositide-dependent kinase (ERK-RSK-PDK) complex is an excellent example to demonstrate how IDRs and SLiMs govern communication between four different kinase catalytic cores to mediate activation and how in molecular terms these promote the formation of kinase heterodimers in a context dependent fashion.
Kinase Domain Cores versus Intrinsically Disordered Linkers
Protein kinases (PKs) are metabolic switches that regulate cellular signaling. They catalyze phosphoryl transfer from ATP to amino acid side chains of their protein substrates. In contrast to metabolic enzymes, which evolved to be efficient catalysts, PKs have evolved to be sensitive molecular switches [1]. In most cases, they are activated by phosphorylation, and then phosphorylate downstream substrate proteins. PKs rarely act alone, as they are part of hierarchically organized cascades or more intertwined kinase networks. Although we know little about how kinases are activated by other kinases, it cannot be overstated how important kinase-kinase complexes are in controlling cellular physiology. We lack structural information on how kinase heterodimers assemble, as these complexes are highly dynamic and are typically short lived.
To understand how one kinase activates another requires that we consider intrinsically disordered regions (IDRs) that typically flank the well-folded kinase cores and small linear motifs (SLiMs) that are embedded in the IDRs [2] (Box 1). This review focuses on the role of disordered PK regions found in different eukaryotic PK families. These are often unique evolutionary additions to ubiquitous kinase domain cores: they provide kinase family-specific allosteric regulatory mechanisms as well as unique combinatorial opportunities for the higher-order assembly of compatible kinase domain cores. We propose that IDRs allow for the functional integration of the kinase catalytic chassis into multienzyme-based regulatory networks, which then collectively control fundamental aspects of biology. Structural snapshots of the interplay between disordered and structured parts of PKs are shown for the extracellular signal-regulated kinase (see Glossary) (ERK)-RSK-PDK complex that controls cell growth. This example demonstrates the communication between different kinases (ERK, PDK1, and RSK) that represent three different kinase families, mitogen-activated PK (MAPK), AGC kinases, and calcium/calmodulin-dependent kinase (CAMK), and highlights the role of IDRs flanking their catalytic cores. We describe first the general features of the kinase core and the IDRs that flank them. We then discuss mechanisms for kinase-kinase core interactions that allow one kinase to phosphorylate another. Finally, we define RSK as a model system where two kinases and multiple IDRs are embedded within a single polypeptide chain. In RSK we can see how IDRs, SLiMs, and phosphoswitches are all engaged in a single protein to orchestrate activation. Using RSK as an example, we provide a general mechanism of action on the role of IDRs in kinases. We advocate that structured kinase domains should be studied in combination with flexible protein regions promoting the formation of higher order complexes and enabling kinase domain cores to be organized into dynamic signaling cascades.
Box 1. IDRs, SLiMs, and Phosphoswitches.
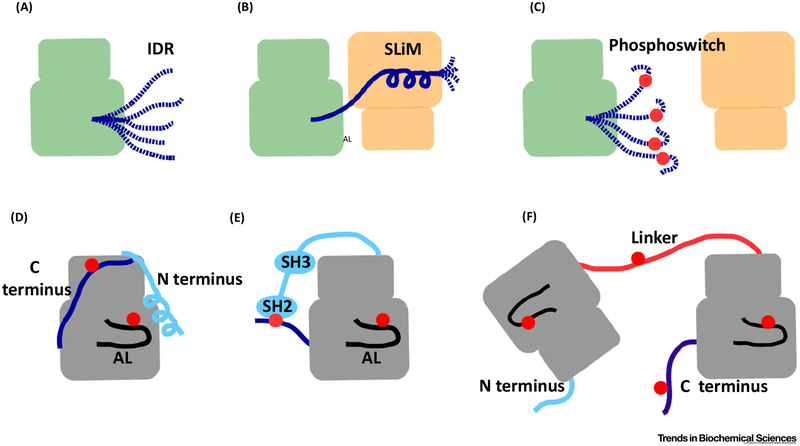
IDRs in proteins do not have a well-defined 3D structure or a stable conformation in solution. Instead, IDRs can adopt multiple conformations. They are ‘fuzzy’, in contrast to polypeptide chains found in structured domains. Because the energy difference in their conformational ensemble is small, perturbations may have a great impact on their structure or order-to-disorder transition. The amino acid composition of IDRs is therefore ideal to sense phosphorylation. Furthermore, IDRs harbor many linear binding motifs and depending on their protein-protein interaction partner, they can mold into different conformation states compatible with the surfaces of their various binding partners [51,52]. In PKs many of the linear binding motifs, frequently referred to as SLiMs, operate as phosphoswitches and harbor phosphorylation target sites for different kinases (Figure I). Phosphorylation of a SLiM can alter its ability to recognize different binding partners. Although SLiMs may affect the activity and protein-protein interactions of kinase domain cores autonomously, it is important to note that they may regulate kinase function through binding to modular protein-protein domains such as SH2, SH3, and 14-3-3.
Figure I. Kinase Domains Are Often Regulated by IDRs.
Schematic representation of IDRs (A), SLiMs (B), and phosphoswitches (C) are shown. (D) N-terminal and C-terminal regions, and the AL are three evolutionarily diverse IDRs that decorate most kinase domain cores. The panel shows these IDRs in the context o fthe protein kinase A kinase domain core. (E) Multi domain kinases possess flexible linkers connecting their kinase domain cores to other regulatory domains. Intervening IDRs connect SH2 and SH3 domains and the kinase domain core of the Src kinase. The C-terminal tail of Src also contains a phosphoswitch. (F) Proteins containing multiple kinase domains such as RSK, in addition to the three IDRs, also possess a fourth IDR, the linker connecting the kinase domains. The linker and the C tail contain SLiMs that function as phosphoswitches. Red dots indicate dynamically regulated phosphoswitches. Abbreviations: AL, activation loop; IDR, intrinsically disordered region; RSK, ribosomal S6 protein kinase; SLiM, small linear motif.
Regulation of Kinase Domain Cores
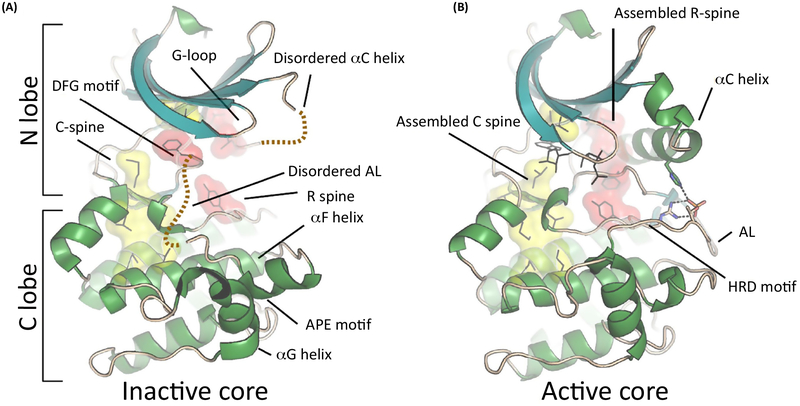
PKs have a bilobal structure, where the N lobe is mainly responsible for ATP binding and the C lobe for substrate recognition. These lobes, which have an independent fold, are connected noncovalently through a central network of hydrophobic amino acids (spines) when the protein is in an active conformation [3]. The catalytic C spine may be regarded as a firm but flexible hydrophobic unit, whereas the N lobe and C lobe components are fused together by the adenine moiety of the bound cofactor (Figure 1). In contrast, the regulatory or R spine, is highly dynamic, and its assembly is required for catalytic activity [4]. The R spine includes a residue from the C helix of the N lobe, and this helix serves as an allosteric hotspot for most PKs. This helix is also responsible for stabilizing a salt bridge between the bound cofactor and a Lys from ß3. In inactive kinases, the C helix is often in an ‘out’ or inactive conformation, or disordered [5].
Figure 1. Common Features of the Protein Kinase Catalytic Chassis.
The two lobes of a prototypical kinase domain are connected through two hydrophobic spines formed by the side chains of its core. (A) In the inactive conformation the spines are not fully assembled and the two kinase lobes are structurally not well connected. (B) Despite that the secondary structural elements are unaffected, the C and R spines assemble only in the ATP-bound active conformation. For most kinases, AL phosphorylation facilitates the assembly of the R spine by affecting the ordering or the positioning of the C helix. Abbreviations: AL, activation loop.
Dynamic IDRs That Drive the Assembly of Active Kinases
IDR I: The Activation Loop Is a Universal Regulator of Kinase Activity
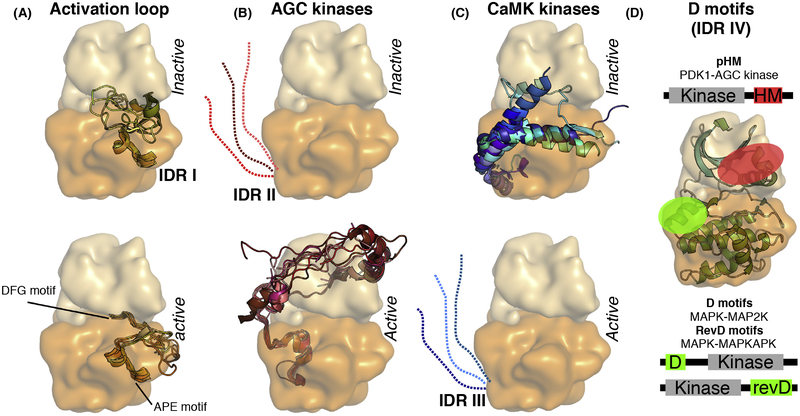
Embedded within the kinase fold is an internal IDR, the activation loop (AL) (Figure 2A). The activation segment starts with the DFG motif and ends with the APE motif. These two flexible sequence motifs are conserved among kinases and have unique functions. The DFG motif lies between the two lobes, right below the C helix and the glycine-rich loop (G loop). Embedded in the DFG motif is another R-spine residue (RS2), which couples directly with the C-helix R-spine residue (RS3) and the catalytic loop R-spine residue [6]. Many kinases display two conformations of this motif: DFG-in and DFG-out [7,8]. In the DFG-out conformation the R spine is disassembled, and in many cases the Phe side chain of this motif faces towards the G loop instead of the C helix. In these cases the DFG-out conformation is thus incompatible with ATP binding (Figure 1A). Interplay between the DFG in/out and the C-Helix in/out is highly regulated and defines the switch mechanism that leads to kinase activation. Both motifs are flexible and must be aligned.
Figure 2. Types of Known Protein Kinase IDRs.
Universal or kinase group specific IDRs often have distinct structural propensity in the inactive versus active state of the kinase. (A) For example, the flexible activation loop of most kinases shows a greater structural propensity in their active versus inactive state (IDR I). (B) Similarly, the C-terminal extension of AGC kinases contains SLiM (pHM), which is more flexible in the inactive state but it can interact with the N lobe in a phosphorylation-dependent manner and becomes ordered in the active conformation (IDR II). (C) In contrast, the C tail of CaMK forms a helical extension in the inactive kinase and this blocks the active site (IDR III). In this case, coactivator binding (e.g.,calmodulin) activates the kinase by competing with the inhibitory helix, thus releasing the steric inhibition (panels were made by using the following protein structures from the PDB: 1ATP, 1IRK, 1QCF, 1OPJ, 1ERK, 1IR3, 2ERK, 2OFV, 3MPM, 3DQW for panel A; 1ATP, 1YM7, 2ACX, 2VD5, 2ESM, 3D0E, 3QKM, 3IW4, 3TXO, 3PFQ, 4L45for panel B; 1TKI, 1KWP, 2VZ6, 2W4K, 2YA9, 2W40, 3RNY, 3KN5, 3MFR, 3BHH, 4MAO for panel C). (D) D motifs are short protein regions located in the IDRs of kinase-binding proteins and they bind to dedicated protein-protein interaction surfaces on kinases. Different kinases may have distinct surface grooves for D motif binding. For example, pHM binds to the PIF pocket located on AGC kinases (shown in red), while unphosphorylated HM binds to the active site cleft of an activator kinase (such as the C-terminal kinase of ribosomal S6 protein kinase). In contrast, the N-terminal D motif from the activator kinases (MAP2Ks) and the C-terminal D motif from the downstream substrate kinases (MAPKAPKs) of MAPKs both bind to another docking surface (shown in green). MAPK-binding D motifs may bind the same so-called docking groove on the kinase in two different N-to-C-terminal orientations (D motifs versus reverse D-motifs, RevD). Abbreviations: AGC kinases, protein kinase A, G and C family; CaMK, calcium/calmodulin-dependent kinase; D motif, docking motif; IDR, intrinsically disordered region; MAP2K, MAPK kinase; MAPK, mitogen-activated protein kinase; MAPKAPK, MAPK activated protein kinase; pHM, phosphorylated hydrophobic motif; PIF pocket, PDK1 interacting fragment pocket; SLiM, small linear motif.
The APE motif precedes the F helix in the C lobe. Although it serves as a stable anchor for the flexible AL, multiple crystal structures captured intermolecularly bind APE motifs in kinase homodimers [9], or alternatively, this region is disordered, suggesting that the APE motif is dynamically anchored to the C lobe. The AL between the DFG and the APE motifs is usually 20 30 residues long and is often partially disordered in inactive structures [10]. Although the AL is highly divergent, it usually contains at least one phosphorylation site, which orders and stabilizes the AL and is typically cooperatively coordinated by an Arg from the catalytic HRD motif (from the C lobe), a basic residue in the AL, and a basic residue from the C helix (from the N lobe) (Figure 1B). The H in the HRD motif is the first R-spine residue (RS1).
IDR II: The C Tail of AGC Kinases Drives Assembly of Their Regulatory R Spine
One of the hallmarks of AGC kinases is their unique C-terminal tail [11]. This tail includes several SLiMs, including the so-called hydrophobic motif (HM), as well as several key regulatory phosphorylation sites (Figure 2B). This flexible region, which anchors the C tail to the N Lobe, stabilizes the C helix in its active conformation and also bridges the links between the N and C lobes. In crystal structures of inactive kinases, the C tail is partially disordered, and the R spine is disassembled (as the C helix is usually twisted or partially unfolded). In an extreme case, as in p90RSK when the C tail is disordered and is in an inactive state, the C helix is replaced by an N-terminal IDR and adopts a unique β sheet conformation [12]. Phosphorylation of the HM triggers assembly of the R spine of the AGC kinases [13]. In the activated AGC kinase the phosphorylated HM (pHM) docks into the so-called PDKI-interacting fragment (PIF) pocket [14], which is formed by the upper part of the C helix and ß4 of the N lobe. The phosphate group mediates charge-based interactions with the N lobe and the hydrophobic residues of the HM extends the R spine.
Apart from the HM, the AL also needs to be phosphorylated. PDK1, an AGC master kinase plays a major role in this process [15]. Although PDK1 lacks its own HM, it has a functional PIF pocket. This allows it to interact with the pHM of most other AGC kinases and to phosphorylate them on their AL. PDK1 is not the only AGC kinase without an HM. Aurora kinases also lack this SLiM, and activation requires binding to another protein, Tpx2, which provides the SLiM and stabilizes its R spine [16].
The outer, solvent-exposed face of the C helix is an allosteric hot spot for regulation of the R spine of the kinase core, and its stabilization is a widespread regulatory mechanism [5]. For example, CMGC kinases use two alternative approaches to stabilize the R spine. In the case of cyclin-dependent kinase, the C helix is situated in a flipped-out position in the inactive state while cyclin binding stabilizes it in a more inward, active position [17]. In contrast, an MAPK rom the same family has a constitutively stabilized active conformation.
Here, the C helix is stabilized by a C-terminal helical extension and kinase activity is simply regulated by AL phosphorylation [17]. Additionally, the negatively charged C-terminal extension of the STE-like GLK kinase can interact with the positively charged C helix intermolecularly [9].
IDR III: The C Tail of CAMKs Governs Access to the Active Site
In some cases both spines of a kinase are assembled, and the AL adopts an activated conformation, yet the kinase is still enzymatically inactive. CAMK type kinases are typical examples [18]. Similarly, to AGC kinases, CAMKs also have a unique, family specific C-terminal extension that is classified as an IDR. Although sequentially, it is not as conserved as the AGC kinase tail, the function of all CAMK tails is the same. The purpose of this helical segment, located close to the active site, is to block ATP and/or substrate binding (Figure 2C). In most cases, this autoinhibited conformation is released in the presence of calcium-bound calmodulin, which pulls this segment off, and thus opens access to the active site [19]. In other cases (e.g., RSK), this inhibitory segment is in a dynamic equilibrium between an open/active and closed/inhibited conformation, and AL phosphorylation shifts the equilibrium towards the active population [20].
While the active site of CAMK type domains is blocked by their C tail, kinases from other groups also share similar features. Pseudosubstrate motifs within terminal extensions, or even within the kinase core can be found across the entire kinome [21,22].
IDR IV: Docking Motifs Facilitate Assembly of Kinase Dimers
Activated kinases may phosphorylate their substrates inefficiently if their phosphorylation target motifs bind to their substrate-binding pocket with low affinity. Therefore, many kinases use additional protein-protein contacts to tether the substrate in close proximity to the kinase and thereby increase the phosphorylation rate of their substrates [23]. These types of proximity-enhancing associations are known as docking interactions and are also thought to be indispensable for the assembly of activator kinase and substrate kinase pairs. A good example of this is the interaction of MAP kinase kinase (MAP2K) (activator) with its MAPK substrate (receiver) kinase, where the docking motif is located in an N-terminal IDR of the activator kinase [24]. Similarly to MAP2K-MAPK binding where the interaction depends on a docking motif located in the N-terminal IDR of MAP2K, the MAPK-MAPK-activated PK (MAPKAPK) activator-receiver kinase pair depends on a C-terminal IDR in the MAPKAPK (Figure 2D) [25].
Kinase-Kinase Core Contacts
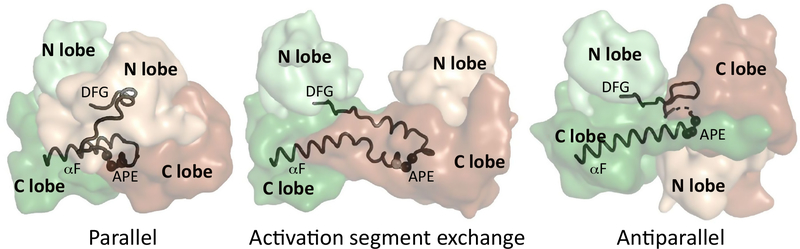
Although the inactive-active transition of a kinase is mediated by various mechanisms often involving IDRs and SLiMs, as discussed earlier, the process naturally also depends on domain-domain contacts between the structured kinase domain core that allows for trans-phosphor-ylation. In order for the phosphorylation sites in the AL of the receiver kinase to gain access to the active site of the activator kinase their structured cores have to make physical contacts. To date, three different ways of kinase dimerization have been described that allow for the AL of the receiver kinase to have access to the active site of the activator kinase (Figure 3). The first involves a dimerization surface on the G helix and shows a parallel kinase dimer arrangement, where both kinases have an N-to-C orientation. This may be referred to as a parallel binding mode. In the complex between KSR2 and MEK1 the two kinase domain cores form a face-to-face, parallel dimer, and a similar interface is observed between the PAK1 homodimers [26,27]. The activation segment exchange model, which is crystallographically captured with various kinase homodimers, also shows a parallel dimer, but here the APE motifs of the two kinases are swapped between the enzyme and the substrate kinase [9,28]. Thus, the unphosphorylated enzyme is thought to phosphorylate its own AL aided by the phosphorylated AL of the partner kinase. The third and more recent model may be referred to as an antiparallel dimer, which reveals a new function of the APE motif [29]. Here, this conserved segment interacts with the G loop of the upstream kinase, exposing the phosphorylation sites of the receiver kinase to the active site of the activator kinase. The CAMK type domain of RSK1 and ERK2 shows a face-to-face, antiparallel dimer, with two main interfaces: the C tail of RSK1 mediates high-affinity binding to the docking surface of ERK, while the APE motif of RSK directly interacts with the cofactor-bound G loop of ERK, bringing the active sites close to each other. It is important to note that these kinase-kinase dimers are distinct from the allosteric activation that is mediated by the back-to-back dimers of BRAF [26,30,31] and the asymmetric dimer of EGFR [32,33]. These latter dimers describe a cis-autophosphorylation mechanism that is distinct from the trans-phosphorylation exemplified by the earlier described face-to-face dimers.
Figure 3. Different Types of Kinase Domain Dimers.
In a parallel hetero dimeric kinase domain complex, the dimerization interface is mainly formed by the G helices of both kinases and the conformation of the APE motif stays as in the kinase monomers. In contrast, in the activation segment exchange model, the activation segments between the upstream and the downstream kinases are swapped; therefore, the APE motif undergoes a major conformational change. In the antiparallel binding mode, the APE motif region also changes its position in order to interact with the ATP-bound G loop of the upstream kinase. This rearrangement is required to expose the rest of the activation loop of the downstream kinase to the active site of the upstream kinase. The activator kinase is colored in brown, the receiver kinase green, and the N and C lobes are shown in light ordarker colors, respectively. The different types of kinase dimers are shown as examples using the following structures from the PDB: parallel-KSR2-MEK1, PDB ID: 2Y4I; activation segment exchange-CHK2, PDB ID: 2CN5; antiparallel-ERK2-RSK1(CTK), PDB ID: 4NIF.
Both the activation segment exchange model and the antiparallel dimer model require a highly dynamic APE motif. In the CAMK type domain of MAPKAPKs, the APE motif indeed shows a lot of flexibility in the inactive state as the C-terminal inhibitory segment displaces the APE motif from its canonical position. This region is either disordered, or it caps an extended F helix similarly as it is observed in the ERK-RSK(C-terminal kinase; CTK) complex [20,29,34–36]. The role of this motif in kinase-kinase dimer formation was previously unappreciated, although it was recognized to be a conserved part of the core kinase fold [37]. While the activation segment exchange theory may explain some aspects of kinase autophosphorylation, its use for heterologous kinase activation is limited, as for the latter, it would require the unlikely assumption that a hybrid enzyme comprising regions from two different kinases would be able to make a functional enzyme with multiple partners. In conclusion, the parallel and the antiparallel dimer-based transactivation modes may be two feasible alternatives to bring the kinase active site from the activator kinase into close proximity to the active site of the receiver kinase. In contrast, the AL exchange mechanism may only be relevant to autoactivation involving homodimers or may turn out to be a crystallographic artifact, given the highly dynamic nature of the APE motifs. To further elucidate this question, we need to obtain structural insights into other heterologous kinase dimers in the future, perhaps from cryo-electron microscopy where there are no crystal packing constraints. We now use RSK as a model system to illustrate how IDRs, SLiMs, and phosphoswitches orchestrate activation.
RSK Activation Requires Interplay between Four Different Kinase Cores
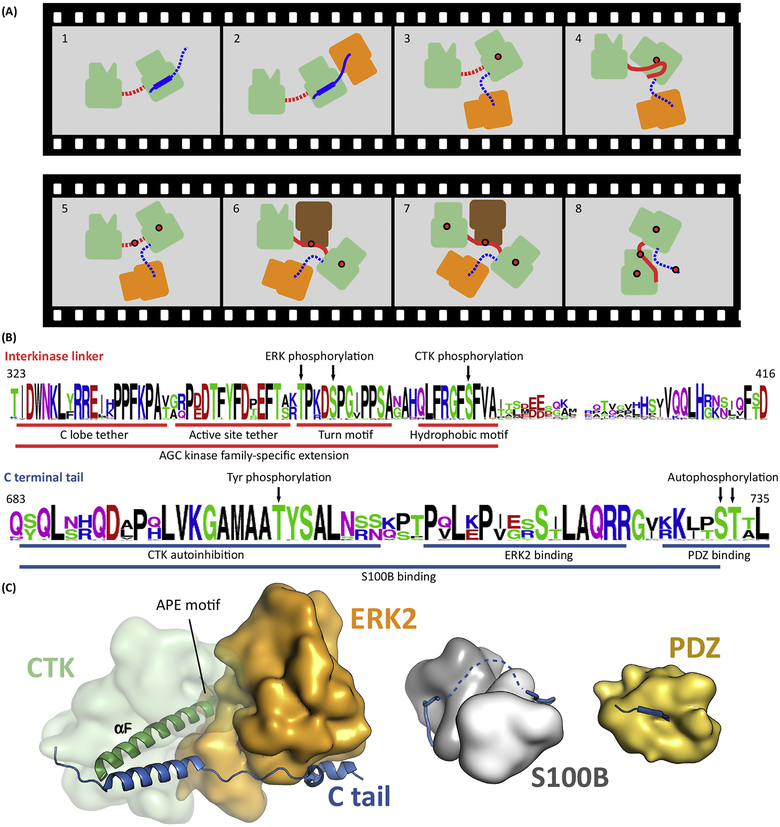
RSK contains an activating kinase (CTK) and the downstream receiver kinase (N-terminal kinase; NTK) in a single polypeptide chain [38], and the initial activating event of ERK binding to the CTK has been captured in a crystal lattice (Box 2). The CTK belongs to the CAMK family, while the NTK belongs to the AGC kinase family [39]. Two additional heterologous kinases, ERK and PDK1, are also required for RSK activation [40]. PDK1 also belongs to the AGC subfamily, so kinases representing three different families are all required for activation of RSK. The NTK and CTK are both flanked by IDRs, and SLiMs that are embedded within these IDRs orchestrate this activation process (Figure 4A). Finally, we can appreciate in RSK how phosphorylation of a SLiM in the linker creates a dynamic phosphoswitch. This interkinase linker shows the characteristic sequence features of IDRs. (i) Low hydrophobicity (namely, low content of bulky hydrophobic amino acids and a high proportion of charged amino acids) apart from a few key positions that mediate some functional aspects (e.g., the AGC kinase family specific extension of this flexible region contains characteristic sequence features important for kinase activity regulation: C-lobe tether, active-site tether, and turn motif; and for protein binding: HM) [11]. (ii) Low evolutionary sequence conservation for those regions that are not involved in of the latter (Figure 4B).
Box 2. IDRs Coordinate Hierarchical Activation of RSK.
RSK contains two kinase domains (NTK and CTK) separated by a linker(Figure I). In addition to their flexible AL, each kinase domain has N-and C-terminal IDRs that play important roles in the hierarchical phosphorylation events initiated by ERK→CTK activation but ultimately leading to the activation of the NTK. NTK activation also depends on the action of a fourth kinase, PDK1. PDK1 is recruited to the multi domain ERK—RSK complex via the linker that contains an HM that functions as a phosphoswitch. The crystal structures of all four kinase domain cores alone have already been determined [20,53–55], but how the kinase domains associate while they phosphorylate each other is structurally not known. The ERK-CTK binary complex crystal structure revealed the role of the C-terminal IDR in holding the complex in a catalytically competent form for the first time [29]. Overall, RSK activation is a good model system to elucidate how different kinase domains work together to orchestrate the activation of the output kinase, in this case the NTK, as well as to shed light on the interplay between structured kinase cores and disordered IDRs that flank the kinase cores.
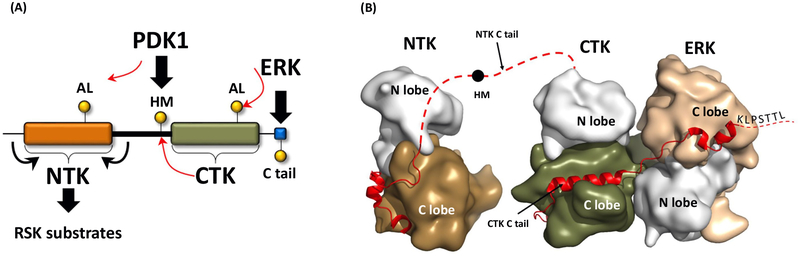
Figure I. Structural Insights into RSK Activation.
(A) Schematic representation of RSK activation. The panel shows the two kinase domains (NTK and CTK), PDK1, and ERK binding SLiMs, and the phosphor switches regulated by the different kinases. (B) Structural model of the ERK-RSK complex. This panel shows the crystal structure of the ERK-CTK binary complex loosely attached to the NTK (dotted red line) and the structure of the inactive NTK. Phosphorylation of the hydrophobic motif(HM)(black dot) in the linker is mediated by the CTK. The Ctail of the CTK is indicated by the red ribbon. Abbreviations: AL, activation loop; CTK, C-terminal kinase; ERK, extracellular signal-regulated kinase; HM, hydrophobic motif; IDR, intrinsically disordered region; NTK, N-terminal kinase; RSK, ribosomal S6 protein kinase.
Figure 4. RSK Requires Coordinated Binding between Four Different Types of Kinase Domains.
(A)Activation of RSK(green) includes several consecutive steps. First, ERK (orange) binds to the C tail of RSK (blue) and activates the CTK by AL phosphorylation (Panels 2 and 3). Activated CTK then phosphorylates the HM located in the linker (red) between the two kinase domains (Panel 4 and 5). Phosphorylated HM recruits PDK1 that phosphorylates the AL of the NTK (Panels 6 and 7). This induces a structural rearrangement in the NTK N lobe, promoting formation of the NTK PIF pocket, allowing binding of the pHM in cis. Ultimately, this last step is required to form the active RSK. In turn, activated RSK phosphorylates the Ctail, which promotes MAPK dissociation. Panel 1: inactive kinase; Panels 2 and 3: CTKAL phosphorylation; Panels 4 and 5: HM phosphorylation; Panels 6 and 7: NTKAL phosphorylation; Panel 8: active kinase. Red dots indicate phosphate groups. Dotted line indicates disordered state. (B) The sequence logo of 40 RSK1/2 orthologs from vertebrates displays the evolutionary sequence conservation of SLiMs (underlined) and phosphoswitches (indicated by arrows) important in Panel A. The upper and lower logos correspond to the linker between the kinases and the C-terminal tail, respectively. (C) Structural models of the MAPK, S100B, and PDZ domain bound RSK1Ctail. Abbreviations: AL, activation loop; CTK, C-terminal kinase domain; ERK, extracellular signal-regulated kinase; HM, hydrophobic motif; MAPK, mitogen-activated protein kinase; NTK, N-terminal kinase domain; pHM, phosphorylated hydrophobic motif; PIF, PDK1-interacting fragment; RSK, ribosomal S6 protein kinase; SLiM, short linear motif.
The C-terminal IDR of the CTK serves as a docking site for the recruitment of several proteins, including the activating ERK. In contrast, the extended linker that joins the two kinase domains of RSK, which is also an IDR, orchestrates the communication between the NTK and CTK. It contains a critical HM that interacts with both kinase domains as well as with PDK1. The HM, a classic SLiM, is first recruited to and phosphorylated by the activated CTK. The pHM then recruits PDK1, which then phosphorylates the NTK on its AL and promotes its activation [41] (Figure 4A,B). The HM is thus a classic phosphoswitch that demonstrates how the functional properties of a SLiM can be dramatically altered by the addition of a single phosphate.
The C-Terminal Tail Is a Multifunctional IDR Controlling Activation of RSK
The 50-amino-acid C tail of the CTK includes many functional motifs (Figure 4B,C). First, it contains an autoinhibitory segment, which is an extension of the CAMK core. Release of this inhibitory segment may be influenced by tyrosine phosphorylation on Tyr707 by FGFR1, for example, which shifts the equilibrium from the autoinhibited to the released position of the inhibitory helix [42]. It also contains a MAPK docking motif, a PDZ binding motif, and Ser/Thr phosphorylation sites [43,44]. The MAPK binding motif is located after the inhibitory segment, and these two motifs are separated by a short linker. The last seven residues of RSK1 also contain an autophosphorylation site that can be phosphorylated by the NTK or the CTK of RSK [45]. This autophosphorylation decreases MAPK binding and thus provides the molecular basis for a negative feedback mechanism [46]. Finally, the C tail includes a PDZ binding motif, which means that this region can also indirectly affect phosphorylation of PDZ-containing RSK substrates by serving as a scaffold that recruits the substrate. These phosphorylation sites are not simply constitutively phosphorylated, but rather play an active role in the dynamics of the response; they are classical examples of phosphoswitches. Phosphatases are also likely to be recruited to these phosphoswitch sites by IDRs. In an alternative scenario, the whole C tail of RSK can bind to S100B [47]. The melanoma marker S100B, which is a homodimer, is an EF-hand-containing protein that resembles calmodulin. S100B can interact with the entire C-terminal IDR of RSK, which dynamically alters the orchestration of RSK regulation [46]. S100B and RSK form a ‘fuzzy’ or intrinsically dynamic complex that affects the catalytic activity of the CTK [48]. In conclusion, the RSK C-terminal IDR directly regulates the enzymatic activity of the CTK [44].
The IDR Connecting the NTK and CTK Governs Communication between the Kinase Domain Cores
The second step in the activation of RSK is the phosphorylation of the intervening IDR that links the two kinase domains (Box 2). This region harbors a classical AGC kinase HM, which is phosphorylated by the activated CTK. The C-terminal tail of the NTK is also embedded within this linker. This includes the HM, which exemplifies a classic phosphoswitch. The unphosphorylated HM is initially recruited to the CTK, but this interaction of the HM with the CTK will be transient. Once the active site of the CTK is open, it will recruit the HM; however, when the unphosphorylated HM docks into the active site of the CTK, it is phosphorylated and then rapidly released. pHM then recruits PDK1 by docking in trans onto the PIF pocket on the C helix of PDK1 [49]. This mediates the formation of another activator-receiver kinase dimer with the NTK. Once PDK1 phosphorylates the AL of the NTK, the pHM most likely docks in c/s to the C helix of the NTK, thus promoting the active conformation of the NTK [49]. In the inactive state, when the HM and ALare not phosphorylated, the NTK has a misfolded and/or disordered N lobe that is incapable of recruiting the linker to assemble its C tail into an active conformation. In contrast, the AL-phosphorylated NTK has a functional PIF pocket with an ordered C helix, which we hypothesize facilitates c/s-interaction with its own pHM. The PDK-mediated phosphorylation of the AL also promotes assembly of the hydrophobic core by stabilizing the active conformation ofthe C helix, while the in c/s binding of the pHM to the NTK PIF pocket extends and thus further stabilizes the R spine. These HM-coordinated, phosphorylation-dependent protein-protein interactions between three different kinases ultimately promote the active conformation of the NTK, which in turn leads to downstream substrate phosphorylation (Figure 4).
Concluding Remarks
Phosphorylation-based regulation is possibly the most widespread regulatory mechanism in cellular signaling. PKs also regulate each other this way. Kinase cascades are an intrinsic feature of these signaling networks. This regulation apparently is not only determined by the common structural elements of the conserved kinase domain chassis, but for many kinases it also involves evolutionarily more divergent IDRs (see Outstanding Questions). It is this IDR-mediated regulation that mostly orchestrates the protein-protein interactions of the kinase cores. Due to lack of insight into full-length kinase structures, as most mechanistic work on kinases have focused on their structured kinase cores, and to the intrinsic dynamic nature of the IDRs, the role of IDRs is understudied and thus not well appreciated. Fortunately, for some kinase families with short IDRs (such as p90RSK), it is becoming apparent that the full functionality of a given PK core can only be understood in the context of the full-length kinase and in its complexes, in particular, with other proteins. RSK serves as an excellent model system for elucidating the role of IDRs in orchestrating the assembly of an active kinase.
Why do PKs form cascades? How do webuild complex switches out of simple catalytic switches, which then can display complex signaling behavior? As we discover more nonlinear behavior in cellular signaling and learn to appreciate the importance of feedbacks (e.g., in setting thresholds, creating binaryswitches, and establishing noise filters) or systems level organization principles, we strive to elucidate the mechanistic basis of all these phenomena [50]. For PK-based signaling activities to be fully understood and appreciated, we believe that it is essential to determine higher-order structures of kinase-kinase complexes. The focus of such mechanistic work needs to be extended to different members of various PK families and must include not only the kinase cores but also the essential IDRs that flank and regulate the cores.
Finally, as PKs control many aspects of cellular life, they are important pharmaceutical drug targets, and more kinase inhibitors will enter into clinical trials. Currently, almost all of these inhibitors target the catalytic activity of the kinases and thus attempt to exploit small differences that exist within the structured kinase domains, even though the specific functions of a PK are mostly underlined by its protein-protein interactions and the higher-order complexes that it makes with other protein partners. This review addresses how IDRs enable kinases to behave as complex molecular switches using p90RSK as a model system. We propose that IDR-mediated interactions may directly be targeted as an alternative to control phosphorylation-based signaling.
Outstanding Questions.
How universal is the dependence of kinase domain cores on additional IDRs? Are there kinase domain associations that do not require flexible contacts but use only rigid parts of their core for dimerization? Is this kinase family dependent?
What are the best methods to determine the structure of full-length kinases containing IDRs? Can we handle the ‘fuzziness’ of unstructured regions with experimental techniques such as cryo-electron microscopy? How can these techniques be bridged with in silico modeling?
How can we modulate the activity of a given kinase with better specificity in a biological context, when it forms a complex with different kinases, for example? How to find drugs that can bind to shallow interfaces characteristic to SLiM mediated binding?
How can we exploit the dynamic nature of kinase-kinase dimerization for the development of new drugs that work based on new mechanistic principles? Are there allosteric sites that may affect the dynamics of binding between kinases? How similar or different are these in different kinase families?
Highlights.
PKs contain IDRs flanking their structured kinase domain cores, which are organized into multienzyme cascades with the help of SLiMs and phosphoswitches.
Because of a common catalytic chassis, there may only be a limited number of ways for activator: receiver kinase complexes to form, albeit this could be promoted and regulated by IDRs in different ways.
p90RSK activation requires the coordinated activation of four different kinase domains where the role of SLiMs and phosphoswitches in the assembly of kinase heterodimers is biochemically well explored.
The full functionality of a given PK core can only be understood in the context of the full-length kinase and its complexes.
IDR-mediated interactions may directly be targeted as an alternative to control phosphorylation-based signaling.
Acknowledgments
This work was supported by the National Research Development and Innovation Office (NKFIH) grants: NN114309 and KKP 126963 (to A.R.) and by the National Insitutes of Health grant DK54441 (to S.S.T.).
Glossary
- Activation loop (AL)
a flexible protein region in kinases, involved in activity regulation by phosphorylation
- APE motif
conserved alanine-proline-glutamate motif that anchors the AL to the kinase domain core,and is implicated in substrate binding
- C helix
an important allosteric regulatory region in all kinases, implicated in ATP binding
- CMGC kinases
a group of kinases named after the initials of some members: CDKs, MAPKs, glycogen synthase kinases, and CDK-like kinases
- DFG motif
conserved aspartate-phenylalanine-glycine motif, activation loop anchor, implicated in ATP binding
- Docking motif
a short linear binding motif mediating binding to PKs
- Extracellular signal-regulated kinase (ERK)
a cell-growth-promoting MAPK
- G-loop
glycine-rich loop, implicated in ATP binding
- HRD motif
conserved histidine-arginine-aspartate motif of the kinase domain core, implicated in catalysis
- Hydrophobic motif (HM)
ahallmark regulatory site for AGC kinases
- PDK1-interacting fragment (PIF) pocket
involved in phosphorylated HM binding
- Phosphoswitch
a linear motif that changes its binding properties upon phosphorylation
References
- 1.Taylor SS and Kornev AP (2011) Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci 36, 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright PE and Dyson HJ (2015) Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol 16, 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kornev AP et al. (2008) A helix scaffold for the assembly of activeprotein kinases. Proc. Natì. Acad. Sci. U. S. A 105,14377–14382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J et al. (2017) A dynamic hydrophobic core orchestrates allostery in protein kinases. Sci. Adv 3, e1600663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor SS et al. (2015) Integration of signaling in the kinome: architecture and regulation of the alphaC Helix. Biochim. Biophys. Acta 1854, 1567–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meharena HS et al. (2013) Deciphering the structural basis of eukaryotic protein kinase regulation. PLoS Biol 11, e1001680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hari SB et al. (2016) Sequence determinants of a specific inactive protein kinase conformation. Chem. Biol 20, 806–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mobitz H (2015) The ABC of protein kinase conformations. Biochim. Biophys. Acta 1854, 1555–1566 [DOI] [PubMed] [Google Scholar]
- 9.Marcotte D et al. (2017) Germinal-center kinase-like kinase co crystal structure reveals a swapped activation loop and C-terminal extension. Protein Sci 26, 152–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nolen B et al. (2004) Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 15, 661–675 [DOI] [PubMed] [Google Scholar]
- 11.Kannan N et al. (2007) The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc. Natl. Acad. Sci. U. S. A 104, 1272–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derewenda U et al. (2013) Identification of quercitrin as an inhibitor of the p90 S6 ribosomal kinase (RSK): structure of its complex with the N-terminal domain of RSK2 at 1.8 A resolution. Acta Crystallogr. D. Biol. Crystallogr 69, 266–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearce LR et al. (2010) The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol 11, 9–22 [DOI] [PubMed] [Google Scholar]
- 14.Biondi RM et al. (2000) Identification of a pocket in the PDK1 kinase domain that interacts with PIF and the C-terminal residues of PKA. EMBO J 19, 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mora A et al. (2004) PDK1, the master regulator of AGC kinase signal transduction. Semin. Cell Dev. Biol 15, 161–170 [DOI] [PubMed] [Google Scholar]
- 16.Cyphers S et al. (2017) A water-mediated allosteric network governs activation of aurora kinase A. Nat. Chem. Biol 13, 402–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson EE et al. (2009) Comparative surface geometry of the protein kinase family. Protein Sci 18, 2016–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Temmerman K et al. (2013) Structural and functional diversity in the activity and regulation of DAPK-related protein kinases. FEBS J 280, 5533–5550 [DOI] [PubMed] [Google Scholar]
- 19.Rellos P et al. (2010) Structure of the CaMKIIdelta/calmodulin complex reveals the molecular mechanism of CaMKII kinase activation. PLoS Biol 8, e1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malakhova M et al. (2008) Structural basis for activation of the autoinhibitory C-terminal kinase domain of p90 RSK2. Nat. Struct. Mol. Biol 15, 112–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ha BH et al. (2012) Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate. Proc. Natl. Acad. Sci. U. S. A 109, 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gogl G et al. (2015) The Structure of an NDR/LATS kinase-Mob complex reveals a novel kinase-coactivator system and substrate docking mechanism. PLoS Biol 13, e1002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller CJ and Turk BE (2018) Homing in: mechanisms of substrate targeting by protein kinases. Trends Biochem. Sci 43, 380–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeke A et al. (2015) Systematic discovery of linear binding motifs targeting an ancient protein interaction surface on MAP kinases. Mol. Syst Biol 11, 837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garai A et al. (2012) Specificity of linear motifs that bind to a common mitogen-activated protein kinase docking groove. Sci. Signal 5, ra74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brennan DF et al. (2011) A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature 472, 366–369 [DOI] [PubMed] [Google Scholar]
- 27.Wang J et al. (2011) Structural insights into the autoactivation mechanism of p21-activated protein kinase. Structure 19, 1752–1761 [DOI] [PubMed] [Google Scholar]
- 28.Oliver AW et al. (2007) Activation segment exchange: a common mechanism of kinase autophosphorylation? Trends Bio-chem. Sci 32, 351–356 [DOI] [PubMed] [Google Scholar]
- 29.Alexa A et al. (2015) Structural assembly of the signaling competent ERK2-RSK1 heterodimeric protein kinase complex. Proc. Natl. Acad. Sci. U. S. A 112, 2711–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu J et al. (2013) Allosteric activation of functionally asymmetric RAF kinase dimers. Cell 154, 1036–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freeman AK et al. (2013) Effects of Raf dimerization and its inhibition on normal and disease-associated Raf signaling. Mol. Cell 49, 751–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X et al. (2006) An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125, 1137–1149 [DOI] [PubMed] [Google Scholar]
- 33.Lemmon MA et al. (2014) The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol 6, a020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng W et al. (2002) Structure of mitogen-activated protein kinase-activated protein (MAPKAP) kinase 2 suggests a bifunctional switch that couples kinase activation with nuclear export. J. Biol. Chem 277, 37401–37405 [DOI] [PubMed] [Google Scholar]
- 35.Jauch R et al. (2006) Mitogen-activated protein kinases inter acting kinases are autoinhibited by a reprogrammed activation segment. EMBO J 25, 4020–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malakhova M et al. (2010) The crystal structure of the active form of the C-terminal kinase domain of mitogen-and stress-activated protein kinase 1. J. Mol. Biol 399, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kannan N and Neuwald AF (2005) Did protein kinase regula tory mechanisms evolve through elaboration of a simple structural component? J. Mol. Biol 351, 956–972 [DOI] [PubMed] [Google Scholar]
- 38.Romeo Y et al. (2012) Regulation and function of the RSK family of protein kinases. Biochem. J 441, 553–569 [DOI] [PubMed] [Google Scholar]
- 39.Cargnello M and Roux PP (2011) Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev 75, 50–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Utepbergenov D et al. (2016) Bacterial expression, purification and in vitro phosphorylation of full-length ribosomal S6 kinase 2 (RSK2). PLoS One 11, e0164343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dalby KN et al. (1998) Identification of regulatory phosphorylation sites in mitogen-activated protein kinase (MAPK)-activated protein kinase-1a/p90rsk that are inducible by MAPK. J. Biol. Chem 273, 1496–1505 [DOI] [PubMed] [Google Scholar]
- 42.Hinsby AM et al. (2003) Signaling initiated by overexpression of the fibroblast growth factor receptor-1 investigated by mass spectrometry. Mol. Cell Proteomics 2, 29–36 [DOI] [PubMed] [Google Scholar]
- 43.Thomas GM et al. (2005) Ribosomal S6 kinase 2 interacts with and phosphorylates PDZ domain-containing proteins and regulates AMPA receptor transmission. Proc. Natl. Acad. Sci. U. S. A 102, 15006–15011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gogl G et al. (2018) Dynamic control of RSK complexes by phosphoswitch-based regulation. FEBS J 285, 46–71 [DOI] [PubMed] [Google Scholar]
- 45.Roux PP et al. (2003) Phosphorylation of p90 ribosomal S6 kinase (RSK) regulates extracellular signal-regulated kinase docking and RSK activity. Mol. Cell Biol 23, 4796–4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gogl G et al. (2016) Structural basis of ribosomal S6 kinase 1 (RSK1) inhibition by S100B protein: modulation of the extracellular signal-regulated kinase (ERK) signaling cascade in a calcium dependent way. J. Biol. Chem 291, 11–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartman KG et al. (2014) Complex formation between S100B protein and the p90 ribosomal S6 kinase (RSK) in malignant melanoma is calcium-dependent and inhibits extracellular signal-regulated kinase (ERK)-mediated phosphorylation of RSK. J. Biol. Chem 289, 12886–12895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miskei M et al. (2017) Fuzziness enables context dependence of protein interactions. FEBS Lett 591, 2682–2695 [DOI] [PubMed] [Google Scholar]
- 49.Frodin M et al. (2002) A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydro phobic motif phosphorylation. EMBOJ 21, 5396–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kholodenko BN et al. (2010) Signalling ballet in space and time. Nat. Rev. Mol. Cell Biol 11, 414–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gouw M et al. (2018) The eukaryotic linear motif resource −2018 update. Nucleic Acids Res 46, D428–D434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tompa P et al. (2014) A million peptide motifs for the molecular biologist. Mol. Cell 55, 161–169 [DOI] [PubMed] [Google Scholar]
- 53.Biondi RM et al. (2002) High resolution crystal structure of the human PDK1 catalytic domain defines the regulatory phospho-peptide docking site. EMBO J 21, 4219–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang F et al. (1994) Atomic structure of the MAP kinase ERK2 at 2.3 A resolution. Nature 367, 704–711 [DOI] [PubMed] [Google Scholar]
- 55.Malakhova M et al. (2009) Structural diversity of the active N-terminal kinase domain of p90 ribosomal S6 kinase 2. PLoS One 4, e8044. [DOI] [PMC free article] [PubMed] [Google Scholar]