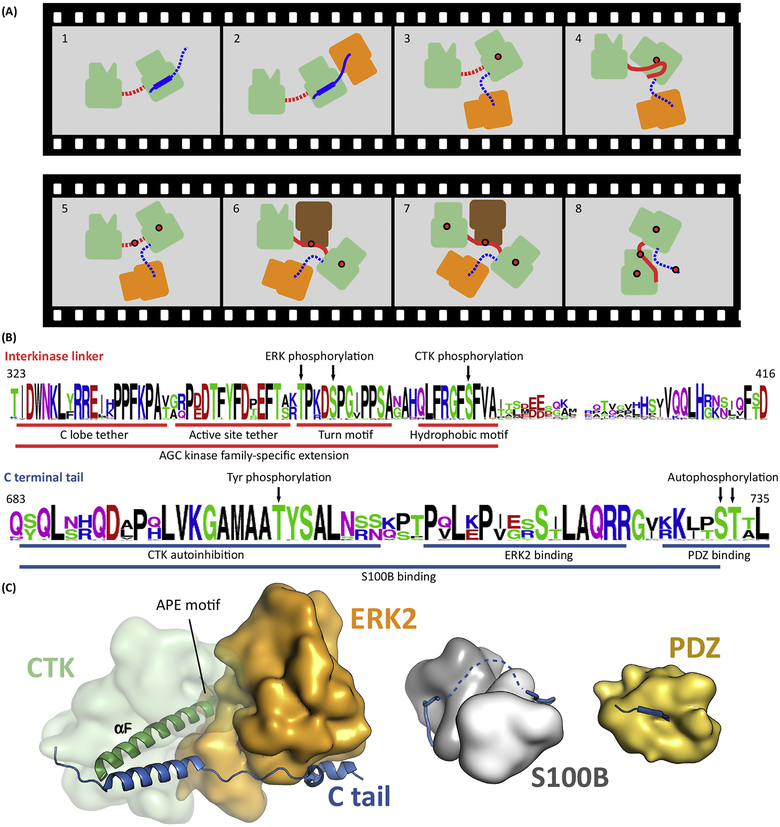
Figure 4. RSK Requires Coordinated Binding between Four Different Types of Kinase Domains.
(A)Activation of RSK(green) includes several consecutive steps. First, ERK (orange) binds to the C tail of RSK (blue) and activates the CTK by AL phosphorylation (Panels 2 and 3). Activated CTK then phosphorylates the HM located in the linker (red) between the two kinase domains (Panel 4 and 5). Phosphorylated HM recruits PDK1 that phosphorylates the AL of the NTK (Panels 6 and 7). This induces a structural rearrangement in the NTK N lobe, promoting formation of the NTK PIF pocket, allowing binding of the pHM in cis. Ultimately, this last step is required to form the active RSK. In turn, activated RSK phosphorylates the Ctail, which promotes MAPK dissociation. Panel 1: inactive kinase; Panels 2 and 3: CTKAL phosphorylation; Panels 4 and 5: HM phosphorylation; Panels 6 and 7: NTKAL phosphorylation; Panel 8: active kinase. Red dots indicate phosphate groups. Dotted line indicates disordered state. (B) The sequence logo of 40 RSK1/2 orthologs from vertebrates displays the evolutionary sequence conservation of SLiMs (underlined) and phosphoswitches (indicated by arrows) important in Panel A. The upper and lower logos correspond to the linker between the kinases and the C-terminal tail, respectively. (C) Structural models of the MAPK, S100B, and PDZ domain bound RSK1Ctail. Abbreviations: AL, activation loop; CTK, C-terminal kinase domain; ERK, extracellular signal-regulated kinase; HM, hydrophobic motif; MAPK, mitogen-activated protein kinase; NTK, N-terminal kinase domain; pHM, phosphorylated hydrophobic motif; PIF, PDK1-interacting fragment; RSK, ribosomal S6 protein kinase; SLiM, short linear motif.