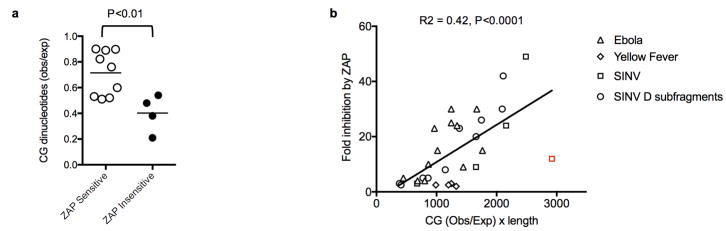
Extended data Figure 8. Analysis of CG-suppression in previously reported ZAP-sensitive and ZAP-resistant viruses and ZAP-sensitizing elements.
a, CG suppression in RNA and reverse transcribing viruses previously reported to be ZAP sensitive (n=9, open symbols) and ZAP resistant (n=4, filled symbols) 7,17–20. The viruses included in the analysis and their degrees of CG suppression (CG observed/expected) are: ZAP-sensitive: Sinbis virus (0.90), Semliki forest Virus (0.89), Venezuelan equine encephalitis virus (0.76), Ebolavirus (0.60), Hepatitis B virus (0.52), Moloney Murine Leukemia Virus (0.51), Marburg virus (0.53), Alphavirus M1 (0.89), Ross River Virus (0.82); ZAP-insensitive: HIV-1 (0.21), Yellow fever virus (0.38) Vesicular stomatitis virus (0.48) Poliovirus (0.54). The p value was calculated using the students T-test (2-sided, n=9 ZAP sensitive viruses and n=4 ZAP resistant viruses). Influenza virus (CG obs/exp = 0.44) that has been reported to be ZAP-resistant due to the presence of an antagonist24 and ZAP-L sensitive via an entirely distinct protein interaction based mechanism23 was excluded from this analysis. b, Analysis of previous published data on ZAP inhibition of reporter gene expression. Each RNA element derived from the indicated RNA viruses was placed in a 3′UTR of a luciferase reporter plasmid and fold inhibition by coexpressed ZAP is plotted against the product of CG suppression (CG observed/expected) and length for each RNA element. A data point that is a quantitative outlier from the general trend (indicated in red) is from the Sinbis (SINV) genome, but is nevertheless included in the linear regression analysis, P value was calculated using the F-test (2-sided, n=32 data points) Data are from references13 and18.