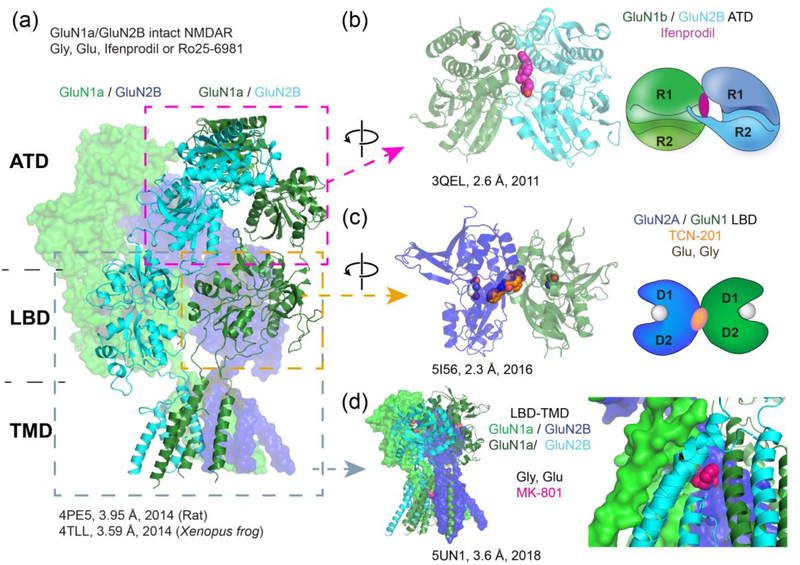
Figure 1. Crystallographic studies on the NMDAR.
(a) Crystal structures of the intact GluN1-GluN2B di-heteromeric NMDARs (without CTD) from rat and Xenopus frog showing a GluN1-GluN2B-GluN1-GluN2B arrangement (GluN1: green/light green, GluN2B: cyan/dark blue) and a domain-swap between the ATD layer and the LBD layer. (b-d) Fragmented approach to effectively monitor ligand binding sites at high resolution. (b) The crystal structure of the GluN1b-GluN2B ATD heterodimers shows the ifenprodil (pink) binding site at the GluN1b-GluN2B subunit interface. Binding of ifenprodil promotes the closure of the bi-lobed structure defined by the upper (R1) and lower (R2) lobes for the GluN2B ATD. (c) A number of crystal structures of NMDAR LBDs, including the ones of GluN1, GluN2A, and GluN3A, have been solved over the years with agonists, competitive antagonists, and allosteric modulators. Shown here is the crystal structure of the GluN1-GluN2A LBDs complexed to glycine (light gray), glutamate (light gray), and TCN-201 (orange). Glutamate and glycine bind at the cleft between the upper (D1) and lower (D2) lobes, whereas TCN-201 binds at the LBD dimer interface. (d) The crystal structure of the GluN1ΔATD – GluN2BΔATD NMDAR shows the binding site of the channel blocker MK-801 (red). Removing the ATDs changes the subunit arrangement in the LBD layer, which highlights the importance of ATDs for proper heterotetrameric subunit assembly. Resolutions of the electron-density maps (in Å) for the respective structures (with PDB code) are stated as in the Protein Data Bank.