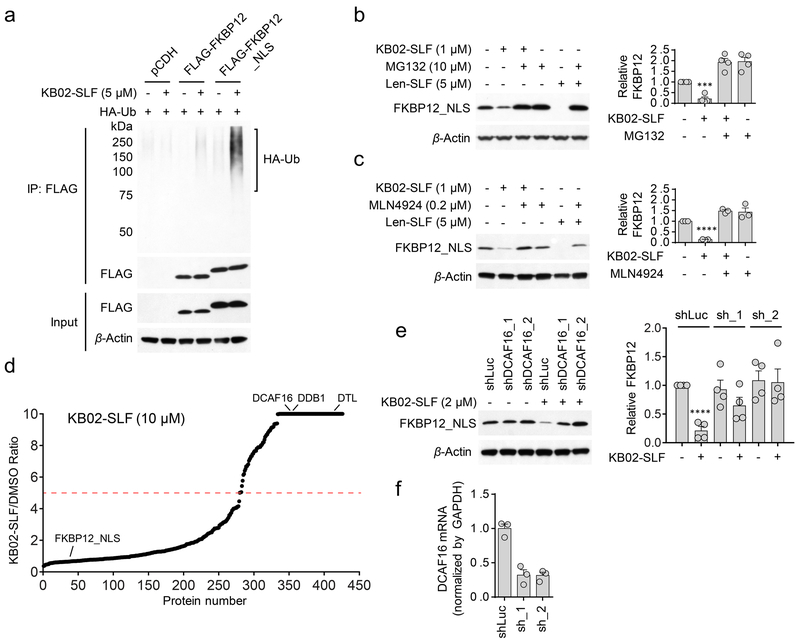
Figure 3. KB02-SLF promotes proteasomal degradation of FKBP12 via the action of Cullin-RING ubiquitin ligases.
a, KB02-SLF mediates polyubiquitination of nuclear (FLAG-FKBP12_NLS), but not cytosolic (FLAG-FKBP12) FKBP12 in HEK293T cells. HEK293T cells stably expressing FLAG-FKBP12 or FLAG-FKBP12_NLS were transiently transfected with HA-Ubiquitin (HA-Ub) for 24 h and then treated with DMSO or KB02-SLF (5 µM) in the presence of the proteasome inhibitor MG132 (10 µM) for 2 h. The result is a representative of three experiments (n = 3 biologically independent experiments). Full images of blots are shown in Supplementary Fig. 14. b, c, KB02-SLF-mediated FLAG-FKBP12_NLS degradation is blocked by MG132 (b) and the neddylation inhibitor MLN4924 (c). HEK293T cells stably expressing FLAG-FKBP12_NLS were co-treated with KB02-SLF (1 µM) and MG132 (10 µM) or MLN4924 (0.2 µM) for 8 h. Lenalidomide (Len)-SLF was used as a positive control. Bar graph (right) represents quantification of the relative FKBP12 protein, with DMSO-treated cells set to a value of 1. Data represent mean values ± SEM (n = 4 biologically independent experiments for (b), n = 3 biologically independent experiments for (c)). Statistical significance was calculated with unpaired two-tailed Student’s t-tests comparing DMSO- to KB02-SLF-treated samples without MG132 or MLN4924. ***P < 0.001; ****P < 0.0001. P values were 0.00016 (b) and 1.8×10−7 (c). Full images of blots are shown in Supplementary Fig. 14. d, SILAC heavy/light (KB02-SLF/DMSO) ratio values of proteins identified in anti-FLAG affinity enrichment experiments (outlined in Supplementary Fig. 5a), where a high ratio indicates proteins selectively enriched from cells treated with KB02-SLF (10 µM). The red dashed line marks a 5-fold ratio value, which was used as a threshold for designating proteins that were substantially enriched by KB02-SLF. The results shown are average ratios from three experiments (n = 3 biologically independent experiments). See method for detailed criteria of data filtering. e, Western blot of stably expressed FLAG-FKBP12_NLS in HEK293T cells transiently transduced with shRNAs targeting DCAF16 (sh_1 and sh_2) or a control shRNA (shLuc) followed by treatment with KB02-SLF (2 µM, 8 h). Right, quantification of the relative FKBP12 protein content, with DMSO-treated cells expressing shLuc set to a value of 1. Data represent mean values ± SEM (n = 4 biologically independent experiments). Statistical significance was calculated with unpaired two-tailed Student’s t-tests comparing DMSO- to KB02-SLF-treated samples. ****P < 0.0001. P value was 0.000036. Full images of blots are shown in Supplementary Fig. 14. f, DCAF16 mRNA was measured by qPCR. Data represent mean values ± SEM (n = 3 biologically independent experiments).