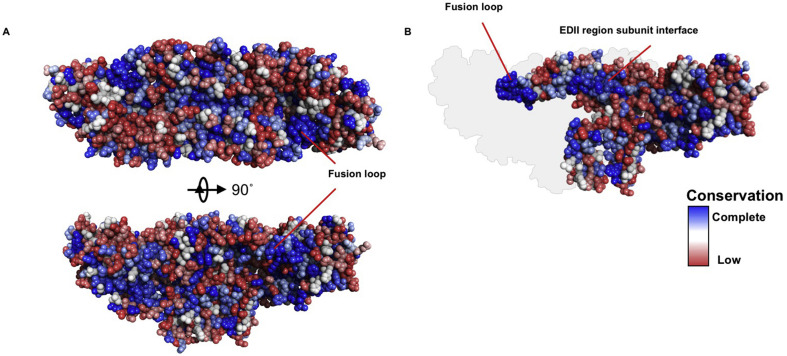
Fig. 1.
Conservation of ZIKV E glycoprotein sequence mapped to representative flavivirus E glycoprotein sequences. (A) A comparative model of ZIKV E glycoprotein and associated prM peptide was generated with Discovery Studio 4.5 (BIOVIA, San Diego, CA, USA). A- CLUSTAL W alignment of representative ZIKV, DENV1, DENV2, DENV3, DENV4, YFV, WNV, JEV, and TBEV E glycoprotein sequences was performed and the relative conservation of residues was mapped to the model. Blue indicates complete conservation among the sequences, shading to white and to red for the least conservation. The ZIKV E glycoprotein dimer is displayed in two orientations. Membrane orientation is behind the protein in the upper panel and below the protein in the lower panel. Conservation is evident in the fusion loop region. (B) ZIKV model as in lower panel of A with one subunit removed to display the interface between the molecules. The deleted subunit is indicated in silhouette. Conservation of the fusion loop and in the subunit interface of the EDII region.