FIG 2.
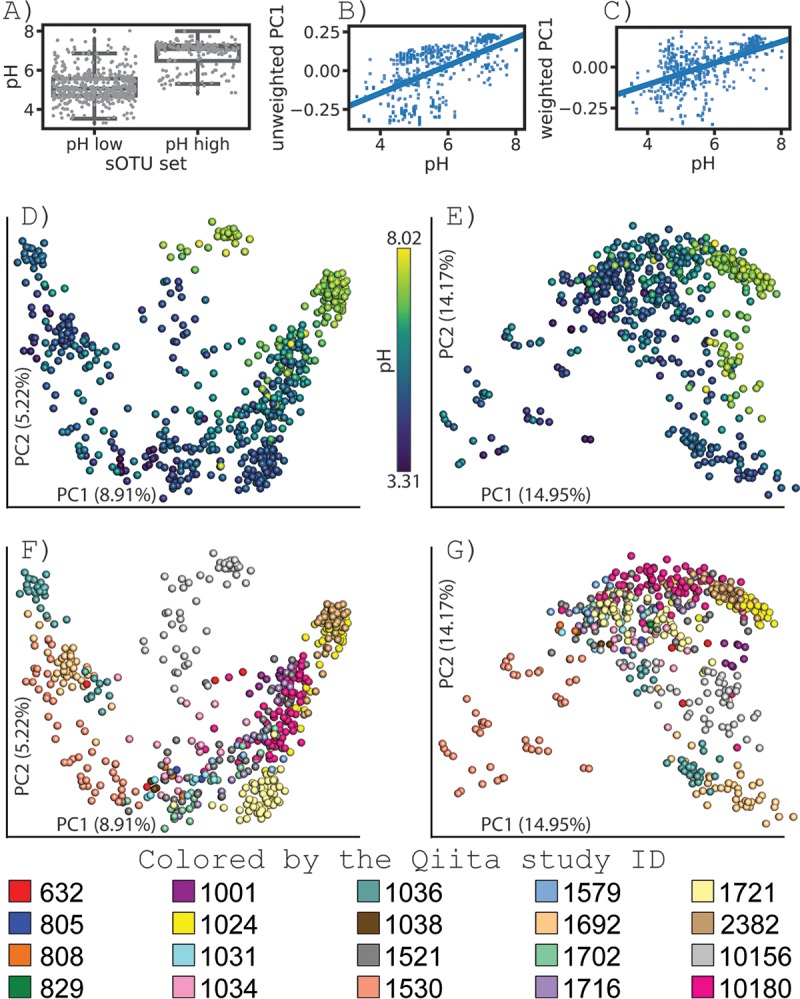
Feature search example. Differential sOTUs from a reanalysis of the study by Ramirez et al. (13) by Morton et al. (unpublished), characterized as associating with a low- or high-pH soil, were obtained. Features were trimmed to 90 nucleotides (nt) to maximize overlap of the Earth Microbiome Project and were searched using redbiom against the Deblur 16S V4 90-nt context with the following sample constraints: “where empo_3=='Soil (non-saline)' and ph > 0.” All samples from Ramirez et al. were removed to create a sample set independent from the observation source: 560 samples remained for assessment following constraints and filtering. (A) Box-whisker plot of the pH values reported in the sample information (Mann-Whitney U statistic = 7,280, P < 9.95 × 10−65). (B and C) Regressions of the reported pH values against the first principal coordinate (PC1) from unweighted (B) and weighted (C) UniFrac analysis (Pearson r = 0.552, P < 6.61 × 10−46, and r = 0.562, P < 6.8 × 10−48, respectively). (D to G) Principal-coordinate plots of unweighted (D) and weighted (E) UniFrac of the observed samples colored by pH and unweighted (F) and weighted (G) UniFrac colored by the Qiita study identifier. (See Table S2 for additional study information.)
