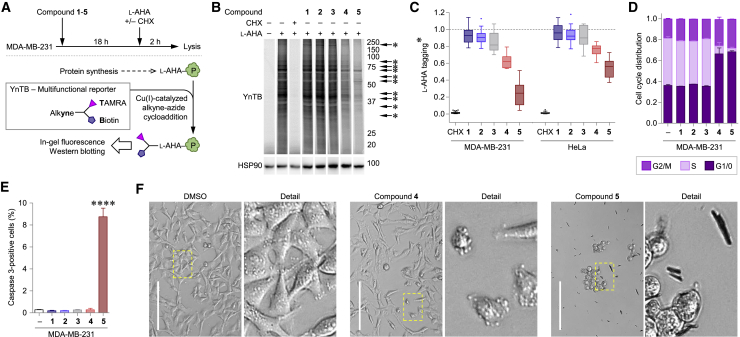
Figure 4.
Effect on Protein Synthesis, Cell Cycle, and Cell Death in MDA-MB-231 Cells
(A) Metabolic tagging of de novo protein synthesis with l-azidohomoalanine (l-AHA). Top: cells pre-incubated with DMSO or 1–5 for 18 h, then pulsed with methionine analog l-AHA for 2 h and lysed. As a positive control for protein synthesis inhibition, cycloheximide (CHX) was applied during the l-AHA pulse. Bottom: l-AHA-containing proteins are ligated by CuAAC to YnTB reporter. Protein synthesis inhibition results in reduced l-AHA incorporation and YnTB reporter fluorescence.
(B) Effects on de novo protein synthesis revealed by l-AHA incorporation. Left to right: cells exposed to DMSO (−) and 1–5. Top: in-gel visualization of l-AHA-tagged proteins. Bottom: loading control HSP90. Full gels are depicted in Figure S2C.
(C) Quantification of l-AHA-tagging in MDA-MB-231 and HeLa cells. Tukey box-and-whisker plot depicts relative fluorescence intensities of n = 10 bands (asterisks in B and Figures S2C and S3C). Plots based on n = 90, 90, 90, 30, 30, 30, 90, 90, 90, 30, 30, and 30 quantifications in duplicate, each individually corrected to loading control HSP90.
(D) Cell-cycle distribution of MDA-MB-231 cells after exposure to 1–5 for 18 h. Cells were analyzed for G2/M, S, and G1/0 through DNA content and proliferation by FACS. For the gating strategy and quantifications, see Figures S5A and S5B, respectively. Mean of n = 3 experiments ± SEM.
(E) Effect on apoptosis. In MDA-MB-231 cells of (D), active caspase-3 protein staining was detected by FACS. For the gating strategy and quantifications, see Figures S5A and S5B, respectively. Mean of n = 3 experiments ± SEM. One-way ANOVA: ****p < 0.0001.
(F) Bright-field micrographs depicting MDA-MB-231 cells exposed to DMSO (left), 4 (middle), and 5 (right) for 18 h. Yellow square depicts the location of the detailed area. Scale bars, 200 μm.