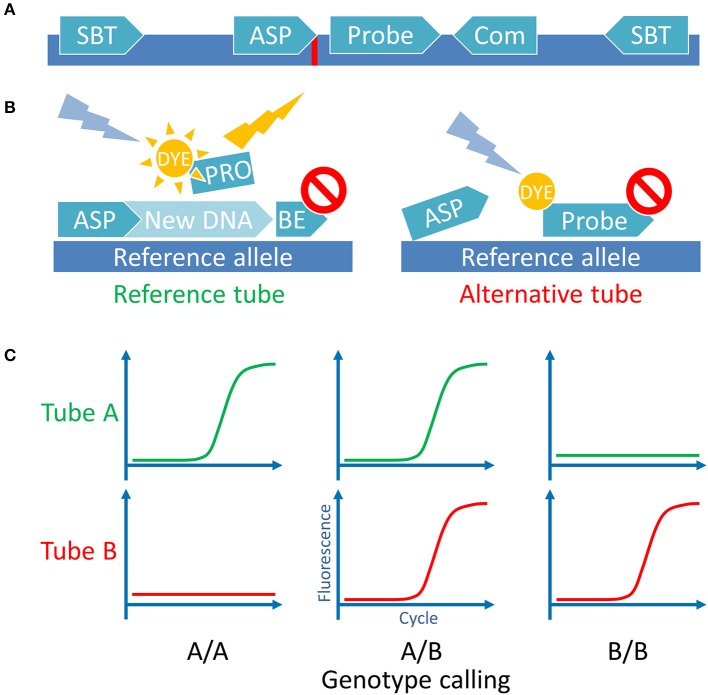
Figure 1.
Assay principle. (A) Schematic representation of the MiHA-coding locus with all the utilized oligonucleotides. SBT, sequence-based typing primers used for Sanger sequencing and for control plasmids cloning; ASP, allele-specific primers used for the AS-qPCR genotyping, complementary with the 3′-end nucleotide to each of the SNP alleles (indicated by red line); Probe, a hydrolysis probe, bearing fluorescent dye and quencher; Com, common primer, used for both SNP alleles AS-qPCRs (for some SNPs common primer was used as the second SBT primer). For UGT2B17/A2 the ASP primers were used as the SBT primers. (B) Schematic representation of the AS-qPCR reaction. The assay is performed in 2 separate tubes with different ASPs for the same SNP. Each tube contains the common gene-specific primer (not shown) and the gene-specific fluorescent probe. Here is represented the genotyping of a DNA sample homozygous for a reference allele. (C) Schematic representation of the possible outcomes of the AS-qPCR and their interpretation. Allele calls are listed below the graphs.