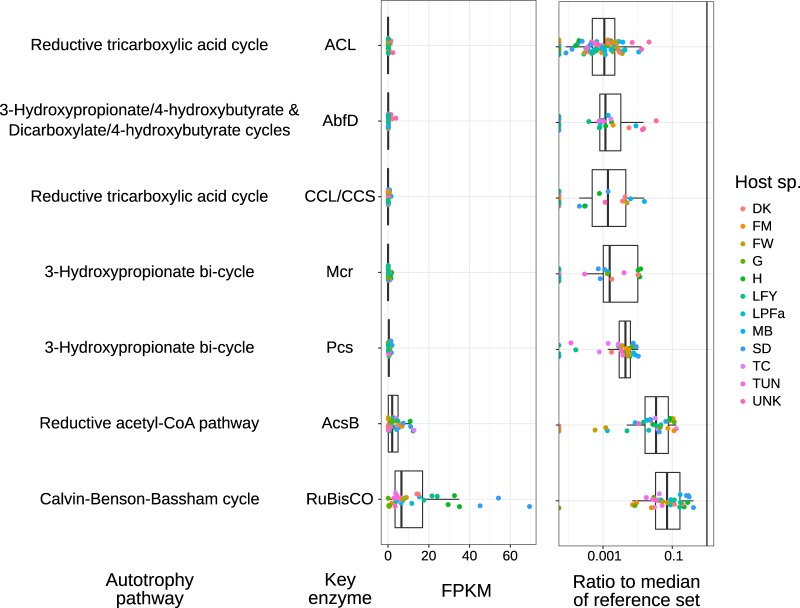
FIG 2.
Read coverage (individual values and box plots) in Kentrophoros metagenomes for key enzymes of autotrophic CO2 fixation pathways, expressed as FPKM values (left) and as a fraction of the median coverage of a reference set of proteins that are expected to be present in all Kentron species (right) (see data posted at https://doi.org/10.5281/zenodo.2575773). Unassembled reads were used for alignment, to rule out the possibility of not detecting hits because of misassembly. Each point represents a separate metagenome library, colored by Kentrophoros host morphospecies. The box midline represents the median, hinges represent the interquartile range (IQR), and whiskers are data within 1.5× IQR of hinges. Abbreviations: ACL, ATP citrate lyase; AbfD, 4-hydroxybutanoyl-CoA dehydratase; CCL/CCS, citryl-CoA lyase/citryl-CoA synthase; Mcr, malonyl-CoA reductase; Pcs, propionyl-CoA synthase; AcsB, CO-methylating acetyl-CoA synthase; RuBisCO, ribulose-1,6-bisphosphate carboxylase/oxygenase.