Figure 1.
ClpB Activation Triggers a Sequential Mode of ATP Hydrolysis
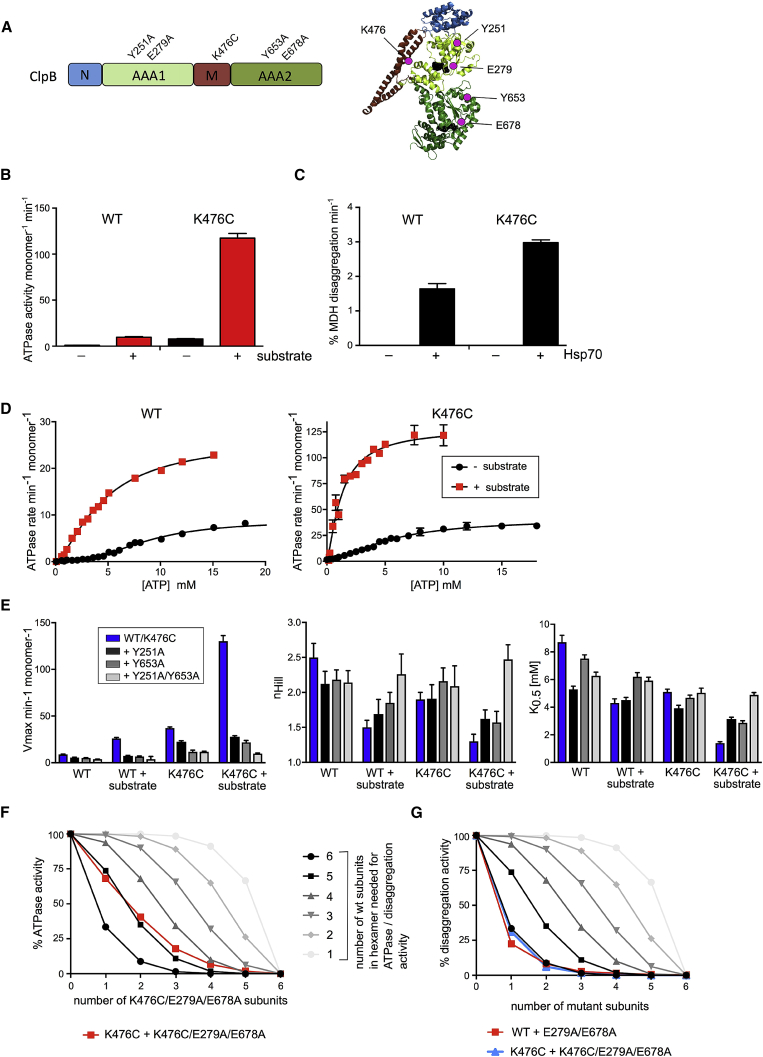
(A) ClpB domain organization and monomer structure. The identity and position of mutated residues are indicated.
(B) ATPase activities of ClpB wild-type (WT) and ClpB-K476C were determined in the absence and presence of 10 μM casein (± substrate). SDs are indicated; for some points, error bars are shorter than the height of the symbol and are not depicted.
(C) MDH disaggregation activities of ClpB-WT and ClpB-K476C in the absence and presence of Hsp70.
(D) ATPase activity of ClpB-WT and ClpB-K476C in absence and presence of casein (± substrate) as a function of ATP concentration.
(E) vmax of ATPase activities, derived Hill coefficient (h), and ATP concentrations at half-maximal ATPase activity (K0.5) for WT, pore 1 (Y251A), and pore 2 (Y653A) loop mutants of ClpB-WT and ClpB-K476C.
(F and G) ATPase activities of ClpB-K476C/ClpB-K476C/E279A/E678A (F) and MDH disaggregation of ClpB-WT/ClpB-E279A/E678A (G) mixes were determined (red, blue). They are compared with curves calculated from a model (black to gray) that assumes that a mixed hexamer only displays ATPase or disaggregation activity if it contains the number of wild-type subunits indicated. Mixing ratios are indicated as number of E279A/E678A mutant subunits.