Extended Data Figure 6 |. Validation of FGF21 binding interface to β-Klotho by ligand binding and cell stimulation experiments.
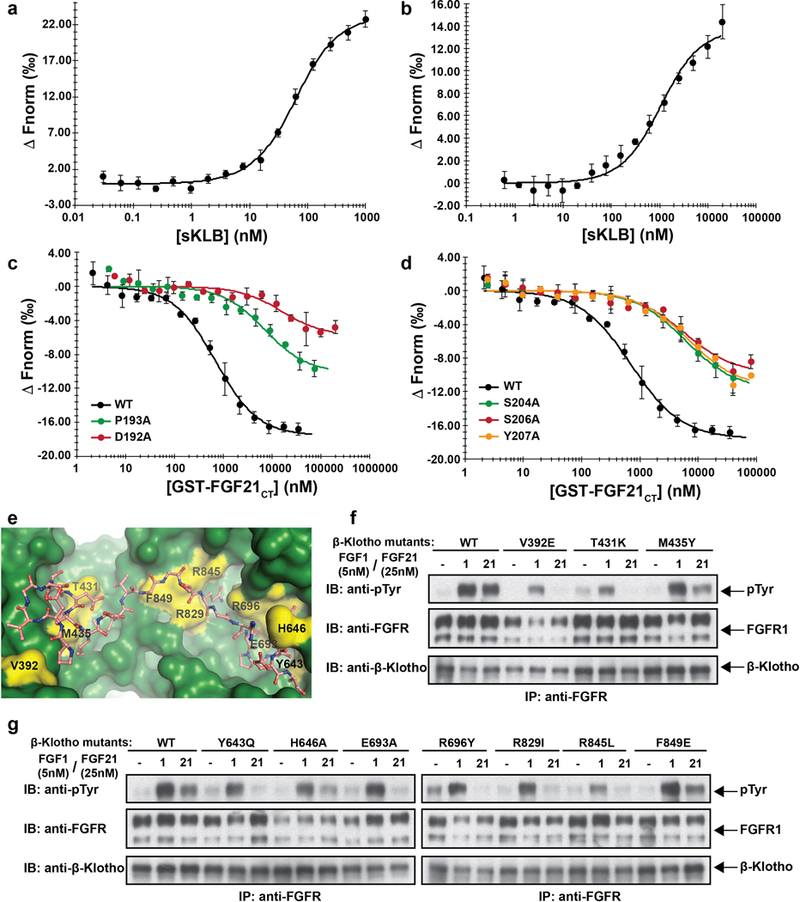
a, b, MST-based binding affinity measurements of (a) FGF21 to sKLB, and (b) FGFR1cD2D3 to sKLB, yielding KD = 43.5 ± 5.0 nM and KD = 940 ± 176 nM, respectively. c, d, MST-based competition assay with GST-FGF21CT containing mutations in either (c) site 1-interacting region or (d) site 2-interacting region. IC50 values for WT, 704 ± 96 nM; D192A, 15900 ± 6210 nM; P193A, 7160 ± 2350 nM; S204A, 5990 ± 1040 nM; S206A, 5560 ± 1590 nM; Y207A, 6630 ± 1570 nM.
The dots and error bars for each graph in panels a-d denote means and variations of ΔFnorm (n = 3 independent samples). Individual experimental data are plotted in Supplementary Fig. 2.
e, Location of mutated amino acid residues (yellow) in sKLB (green) occupied by FGF21 (salmon) that were analyzed in panels f and g. f, g, Stably transfected L6 cells co-expressing FGFR1c together with WT or β-Klotho mutants were stimulated with either FGF21 or FGF1 (control) and analyzed for FGFR1c activation by monitoring tyrosine phosphorylation of FGFR1c. Lysates of ligand stimulated or unstimulated cells were subjected to immunoprecipitation with anti FGFR1 antibodies followed by immunoblotting with either anti-pTyr or anti-FGFR1 antibodies.
