FIGURE 1.
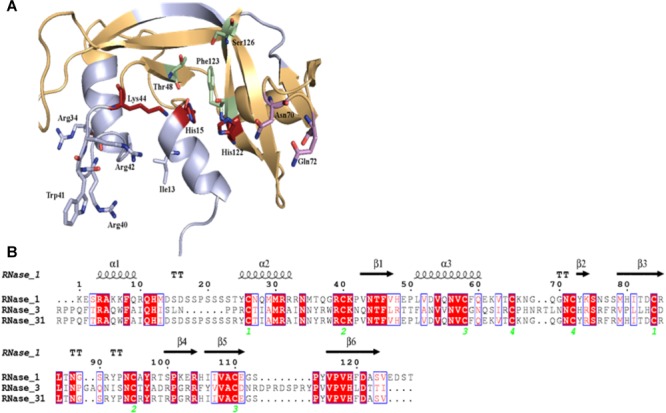
Design of RNase3/1 chimera. (A) Model of RNase 3/1 obtained by Modeler 9.12 (Webb and Sali, 2016). In green, B1 site. In pink, B2 site. In red, the active site. In beige, the RNase 1 skeleton. In light blue, the RNase 3 antimicrobial regions. (B) Alignment of RNase 1, 3 and 3/1 primary structures. The fully conserved amino acids are highlighted in red. The residues that are not conserved but have similar properties are marked with red letters. The secondary structure is indicated above the alignment. The green numbers indicate the disulphide bridge pairs. The alignment was done using Clustal Omega (Sievers and Higgins, 2018), and the image was obtained with ESPript7 (http://espript.ibcp.fr/ESPript/) (Robert and Gouet, 2014).
