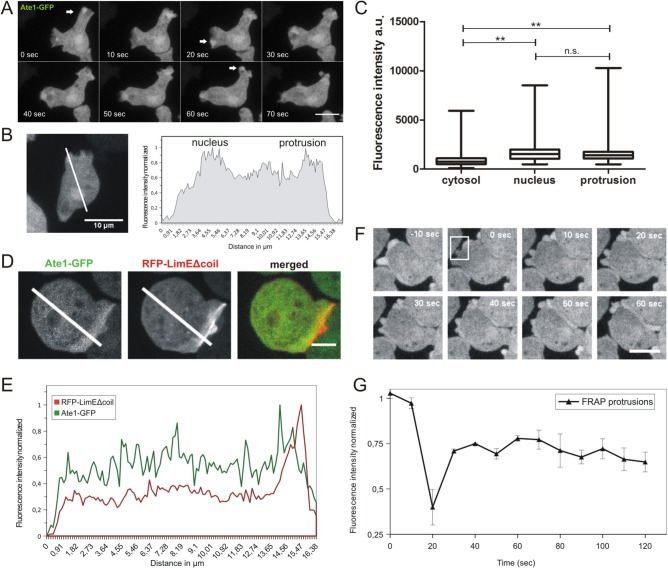
FIGURE 2:
Subcellular localization of DdAte1. (A) DdAte1-GFP expressing cells were recorded by live-cell spinning disk confocal microscopy. DdAte1-GFP localizes to the cytoplasm and is enriched in nuclei and in the cell cortex during the formation of pseudopodia. (B) Live-cell spinning disk confocal microscopy of a DdAte1-GFP expressing cell (left). The intensity profile along the line is plotted (right). The fluorescent signal is stronger in the nucleus and cortical pseudopodia. (C) Fluorescence intensity of DdAte1-GFP in the cytosol, nucleus, and protrusions. Fluorescence intensities of the region of interest were measured with ImageJ within a 4 × 4 square pixel area (0.251 μm2) for n = 54 cells. Fluorescence intensity in protrusions and nuclei showed an enrichment compared with the cytosol. For the statistical analysis, GraphPad Prism Software employing one-way analysis of variance (ANOVA) was used. There is no significance difference between nucleus and protrusion (ns), whereas differences of cytosol versus nucleus and cytosol versus protrusions are significant (p < 0.0097) (**). Bars are ±SD. (D) Live-cell spinning disk confocal microscopy of a Dictyostelium cell expressing both DdAte1-GFP and RFP-LimEΔcoil. (E) The intensity profiles of both fluorescence channels along the lines in D are plotted. (F) Selected time points of a representative FRAP experiment at the protrusion of DdAte1-GFP expressing cells (Supplemental Movie 1). The fluorescence in a protrusion (within the white square at time point 0 s) was bleached with a point-focused 473-nm laser pulse. Spinning disk confocal microscopy images with a time-lapse acquisition rate of six stacks per minute and a maximum intensity projection of five slices per image stack are shown. (G) DdAte1-GFP in protrusions shows a high exchange rate. The graph shows recovery kinetics from three independent measurements performed using a spinning disk microscope (mean ± SD). Scale bars, 5 μm in A, D, and F; 10 µm in B.