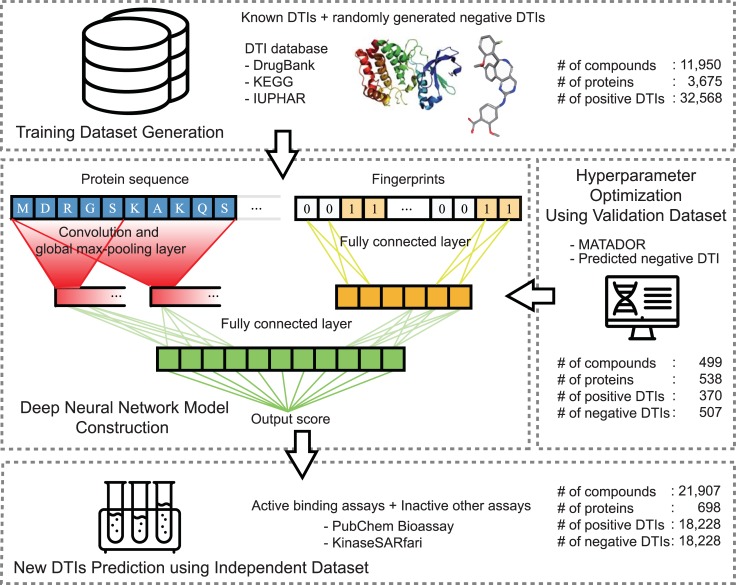
Fig 1. Overview of our model.
First, we collected training DTI datasets from various databases (DrugBank, KEGG, IUPHAR). Second, we constructed the neural network model using convolution, which is able to capture local residue patterns that can help the DTIs. Third, we optimized the hyperparameters with an external validation dataset that we constructed. Finally, we predicted DTIs from bioassays (independent test dataset) and evaluated the performance of our model. The numbers (#) of compounds, proteins and DTIs are summarized in each step.