Figure 3.
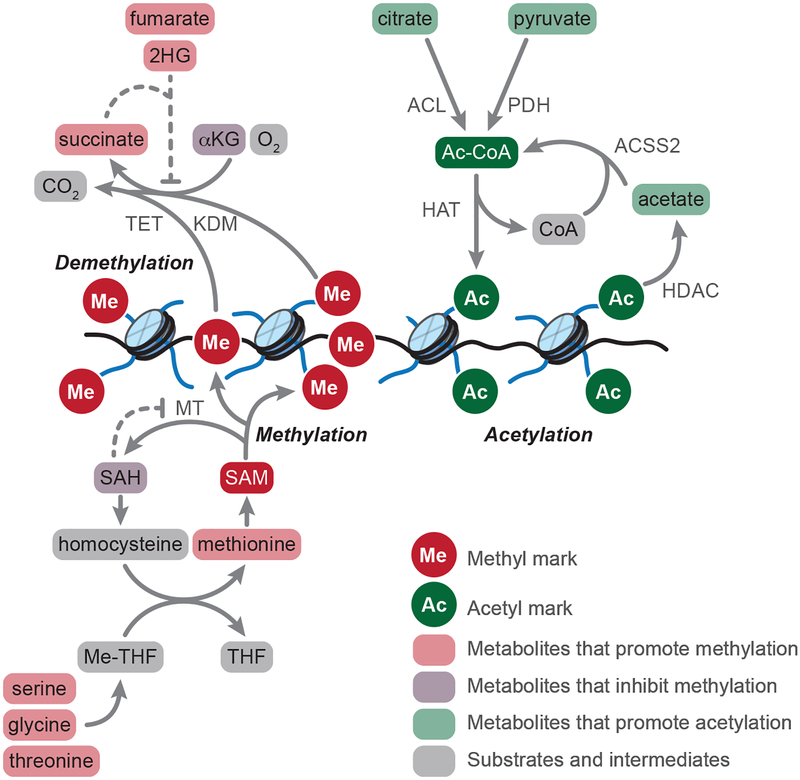
Metabolic regulation of chromatin marks. Methyltransferase (MT) enzymes transfer methyl (Me) groups from S-adenosylmethionine (SAM) to histones and DNA. Methylation reactions generate S-adenosylhomocysteine (SAH) as a product; SAH inhibits methylation reactions, making MT activity responsive to the relative ratio of SAM:SAH. Methionine serves as the direct precursor for SAM, and methionine pools are regulated both by cellular uptake (not shown) and the methionine cycle, which regenerates methionine from homocysteine. The methionine cycle is intimately connected to the folate cycles, in which serine, glycine, and (in mouse PSCs) threonine provide one-carbon units to tetrahydrofolate (THF) for transfer to homocysteine. Lysine demethylases (KDM) and ten-eleven translocation (TET) enzymes catalyze demethylation of histones and DNA, respectively, using alpha-ketoglutarate (αKG), oxygen, and ferrous iron (not shown) as substrates. Succinate, fumarate, and 2-hydroxyglutarate (2HG) function as competitive inhibitors of KDM and TET enzymes, thereby promoting accumulation of methyl marks. Histone acetyltransferases (HAT) transfer acetyl (Ac) groups from acetyl-coenzyme A (Ac-CoA) to histones. Ac-CoA derives from cytosolic and/or nuclear citrate, pyruvate, and acetate, which serve as substrates for the Ac-CoA generating enzymes ATP citrate lyase (ACL), pyruvate dehydrogenase (PDH), or acyl-coenzyme A synthetase short-chain family member 2 (ACSS2). Moreover, class I, II, and IV histone deacetylase (HDAC) reactions generate acetate as a product, which can be captured by nuclear ACSS2 to regenerate Ac-CoA.