Fig. 6. Biological consequence of SENP7 alternative splicing for E2F1 activity.
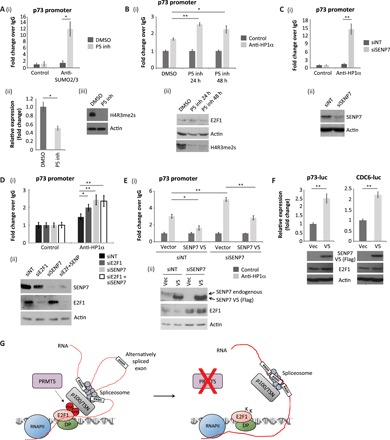
(A) U2OS cells were treated with 5 μM PRMT5 inhibitor for 72 hours, where indicated, before ChIP analysis with anti-SUMO2/3–specific or control antibodies. Immunoprecipitated chromatin was analyzed using primers specific for the E2F site in the p73 promoter (i). An RT-PCR was also performed to monitor the levels of p73 transcripts in the cell (ii). An immunoblot for H4R3me2s is included to demonstrate the activity of the PRMT5 inhibitor (iii). n = 3. See also fig. S4 (F and G). (B) As described above, although cells were treated with the PRMT5 inhibitor for 24 or 48 hours as indicated. ChIP analysis was performed with anti-HP1α–specific or control antibodies (i). An immunoblot for H4R3me2s is included to demonstrate the activity of the PRMT5 inhibitor (ii). n = 2. (C) U2OS cells were transfected with SENP7 siRNA or nontargeting siRNA (siNT) for 96 hours as indicated. Cells were then prepared for ChIP analysis as described above (i). An immunoblot is included to demonstrate input protein levels (ii). n = 4. (D) ChIP analysis as described above, although U2OS cells were transfected with siRNA targeting E2F1, SENP7, or a combination of the two (siE2F1 + siSENP7). n = 3. (E) U2OS cells were transfected with siRNA targeting SENP7 or nontargeting siRNA for 96 hours, as indicated. Cells were subsequently transfected for 48 hours with an empty vector or a plasmid expressing Flag-tagged SENP7 V5. Cells were then prepared for ChIP analysis as described above (i). An immunoblot is included to demonstrate input protein levels (ii). n = 3. (F) U2OS cells were transfected with p73–luciferase (luc) or CDC6-luciferase reporter plasmids for 48 hours, along with empty vector (vec) or Flag-tagged SENP7 V5. Reporter activity was measured, and immunoblots were performed to monitor input protein levels. n = 2. (G) Model diagram where PRMT5-mediated methylation of chromatin-associated E2F1 mediates its interaction with p100/TSN, which permits the E2F1 complex to associate with a subset of RNAs, some being derived from E2F-target genes. By regulating the activity of the splicing machinery, it is proposed that the E2F1-p100/TSN complex can influence the alternative splicing of these RNAs. In the absence of E2F1 methylation (either under conditions of PRMT5 inhibitor treatment or in cells expressing E2F1-meR point mutants), a p100/TSN-dependent interaction with the splicing machinery is lost, and changes to alternative splicing of a subset of RNAs result.
