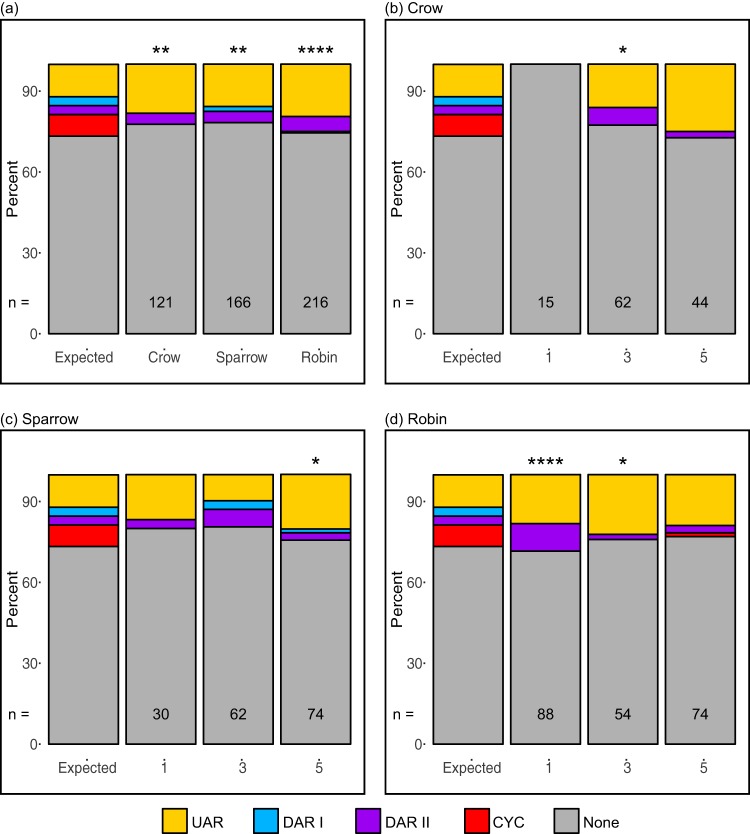
FIG 5.
The frequency of variants within designated binding sites in the WNV 5′ UTR differed from the expected distribution. (a) When the variants were pooled by passage (numbers 1, 3, and 5 on the bottom of each graph), all bird-passaged WNV populations had fewer variants than expected in the CYC binding site. (b to d) Variants located in binding site sequences by passage for viruses passaged in crows (b), sparrows (c), and robins (d) were less common in all binding sites except the UAR. Full statistics from χ2 tests are in Table S5. The UFS riboswitch was also analyzed, and results are described in the text; however, as this sequence overlaps binding sequences, it could not be illustrated in this figure. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.