Fig. 7.
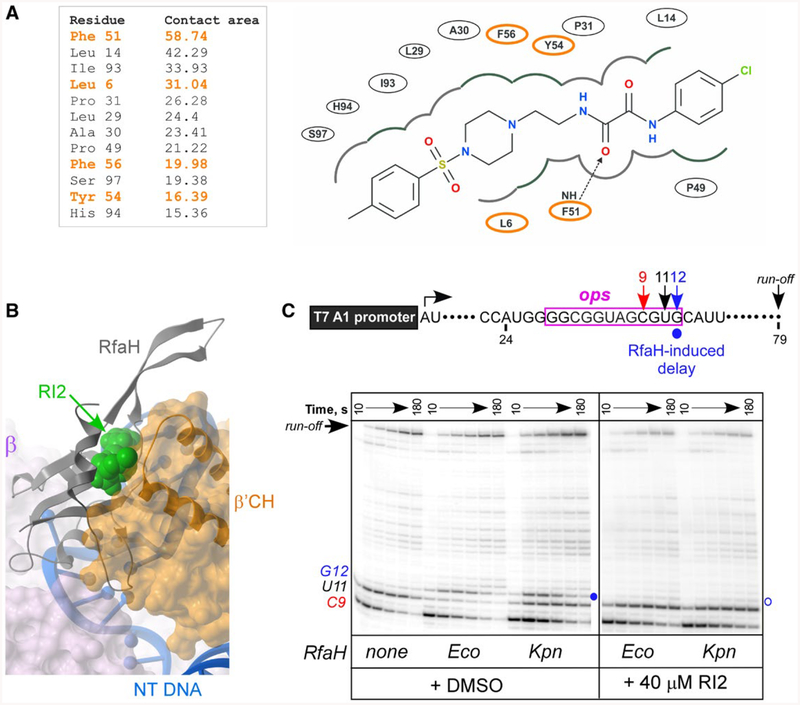
RI2 is predicted to block RfaH interactions with the β’CH domain. A. RfaH residues interacting with RI2 in the predicted binding pose. Residues indicated in orange interact with β’CH of RNAP (Kang et al., 2018). Left, contact areas (Å2) of the RfaH-NTD residues interacting with RI2. Right, a 2D interaction diagram of RfaH-NTD and RI2 in the predicted model. The dashed line with an arrow represents a hydrogen bond between residues and RI2.
B. Superposition of the modeled RfaH-NTD/RI2 complex and the cryo-electron microscopy structure (PDB ID: 6C6T) of the RfaH/TEC complex (Kang et al., 2018) using the RfaH-NTD backbone. Binding of RI2 (green) is incompatible with the β’CH (orange).
C. Effects of RI2 on RfaH recruitment at the ops site. Halted A24 TECs were formed as described in Materials and Methods. Elongation was restarted upon addition of NTPs and rifapentin in the presence of Eco or Kpn RfaH (100 nM) and RI2 (or DMSO). Aliquots were withdrawn at selected times and analyzed on a 10% denaturing gel. Positions of the paused and run-off transcripts are indicated; the position of the RfaH-induced RNAP pause at G12 is indicated with a circle.