Figure 1. Schematic overview of histone cluster deletion and molecular verification.
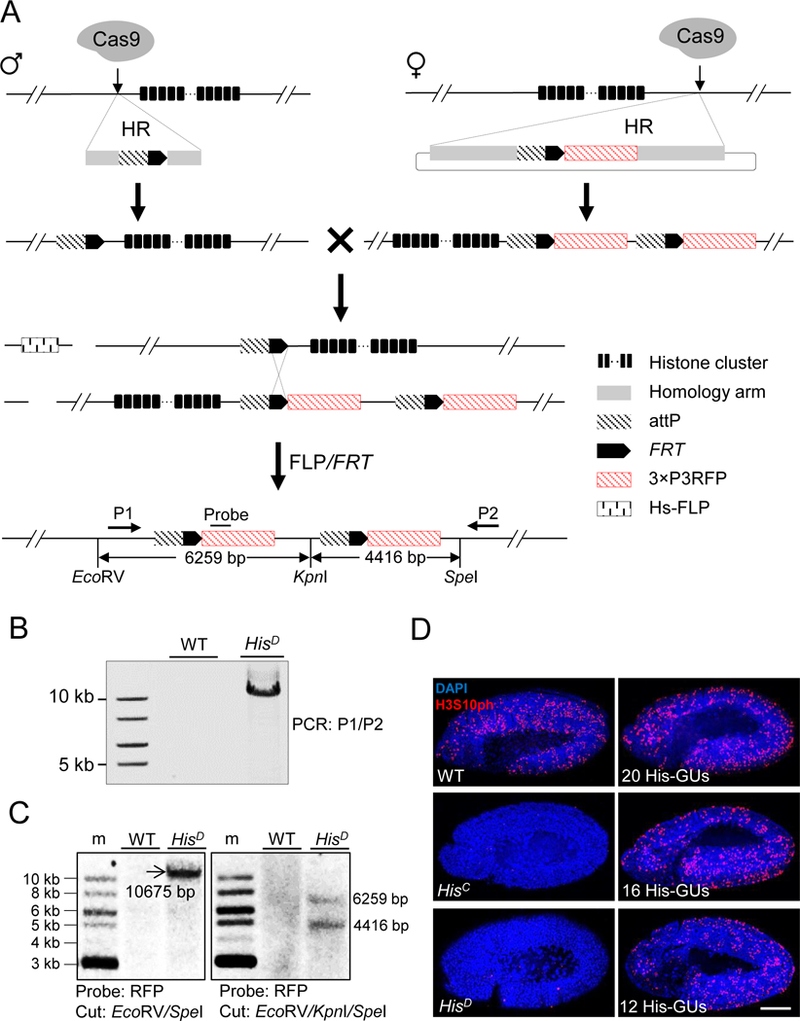
(A) Strategy for knocking out the entire histone cluster. CRISPR/Cas9-mediated homologous recombination (HR) pathway was used to knock-in attP-FRT cassette on both sides of the histone cluster. On the left side, single-stranded oligonucleotide DNA was used as the HR donor. On the right side, a plasmid with homologous arms was used. Flies with ‘ends-in’ HR events were identified during knocking-in at the right side, creating an attP-FRT duplication (top). The two fly lines were crossed and flippase activity was induced. Through HR between FRT sequences (middle), a histone null mutant was generated (bottom). Primers (P1 and P2) for long-range PCR, probe and restriction-enzyme sites for Southern blotting are indicated. The distances between these restriction sites are labeled.
(B) PCR analysis with P1 and P2 primers was used to validate histone cluster deletion (HisD). w1118 wild-type (WT) flies were used as a control.
(C) Southern-blot analysis of HisD. Size markers (m). Genomic DNA from either WT (w1118) or HisD was digested with EcoRV/SpeI (Left) or EcoRV/KpnI/SpeI (Right), separated and blotted with the probe indicated in Figure 1A.
(D) Confocal images of cycle 15 embryos immunostained with H3S10ph antibody (red) and DAPI (blue). WT: w1118; HisC: homozygous histone deletion (Bloomington Stock Center 8670); HisD: homozygous histone cluster deletion (generated in this study); 20His-GUs, 16 His-GUs, and 12His-GUs indicate homozygous histone cluster deletions with different copy numbers of histone gene units (His-GUs). No H3S10ph signal was detected in HisD and HisC embryos. Scale bar: 100 µm.
See also Figure S1.
