Figure 4. Systematic mutagenesis of Drosophila melanogaster histone H3 and H4 residues.
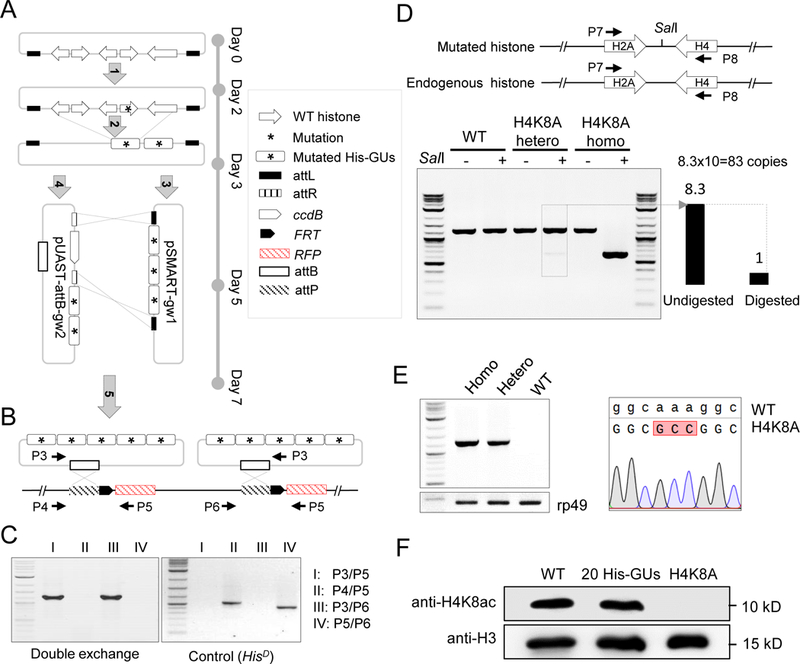
(A) Schematic representation of histone-mutagenesis procedures with Gateway assembly system (see details in Methods). ‘Five-step’ assembly was performed to insert five mutated-histone gene units (His-GUs) into the pUAST-attB integration vector.
(B) PhiC31-mediated ‘attB/attP’ exchange system for integration of two mutated 5 His-GUs into the genome in the HisD fly. Primer pairs for PCR verification are indicated.
(C) PCR verification of double attB/attP recombination.
(D) Molecular characterization of H4K8A mutant flies. The top panel is the schematic representation of the synthetic and native histone genes. Primers to amplify part of the histone genes are indicated. The SalI site introduced in the synthetic construct is shown. The bottom panel shows the gel image of amplified PCR fragments with or without SalI digestion. The copy number of His-GU was estimated as the ratio of band intensity of digested and undigested fragments, and is shown at the bottom left.
(E) Sequence confirmation of the H4K8A mutant. The left panel shows the PCR result using primers specifically to amplify the mutated DNA. The rp49 gene was used as a loading control. The sequencing trace is shown on the right.
(F) Analysis of H4K8ac (acetylated H4K8) in different fly lines by western blot.
See also Figure S4.
