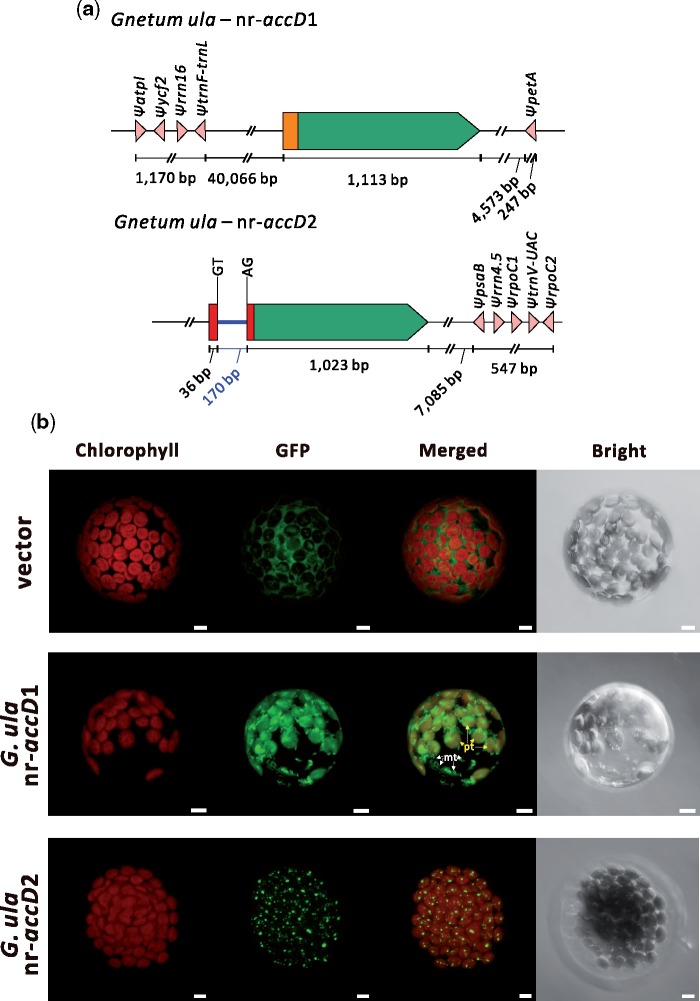
Fig. 4.
—Two nr-accD genes in Gnetum ula have distinct architectures and targeting sites. (a) The nr-accD gene architectures in the G. ula draft genome. The green box and blue line denote the exon and intron, respectively. Orange and red boxes correspond to the distinct TPs of the two nr-accDs in G. ula. Sequences with high similarity to G. ula plastid genes are designated as pink triangles. The intron donor and splice sites are labeled with GT and AG, respectively. Figures are not drawn to scale. (b) Transient expression assays using nr-accD TPs of G. ula indicate that the nr-accD1 dually targets plastids and putative mitochondria, but nr-accD2 likely targets plastoglobuli. Red and green signals in the first two columns represent chlorophyll autofluorescence and GFP, respectively. “Merged” column shows combined chlorophyll and GFP signals, whereas “bright” indicates bright field picture of the protoplasts. The yellow and white arrows in G. ula nr-accD1 merged column indicate plastids and mitochondria localization, respectively. Scale bar = 5 µm. Pt, plastid; mt, mitochondria.