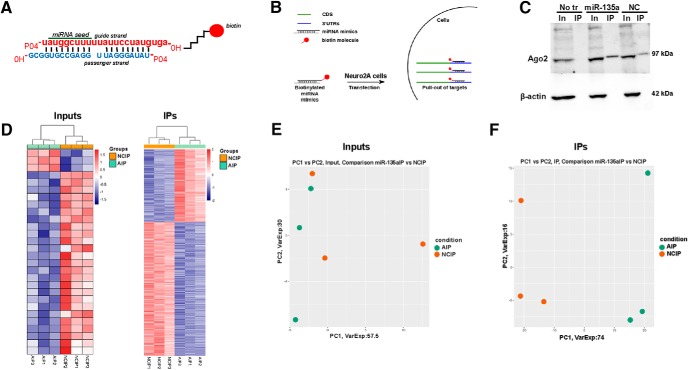
Figure 4.
Target identification for miR-135a using biotinylated probes. A, Schematic of miRNA duplex design. The mature strand is labeled with a biotin molecule at the 3′ hydroxyl group via a C6 linker. B, Schematic showing the IP procedure. Neuro2A cells were transfected with biotin-tagged probes. IP was performed using streptavidin beads. Total RNA was extracted and subjected to deep RNA sequencing. n = 3 biological replicates/group. C, Representative Western blot for Ago2 in miR-135a and negative control IP samples. β-actin was used as a loading control and only present in input samples. No tr, No transfection control; NC, negative control; In, inputs; IP, immunoprecipitates. D, Heatmaps of RNA from input and IP samples showing differential gene expression. E, F, Principal component analysis plots of inputs and IPs showing the clustering of samples basing on their differential gene expression. NCIP, Negative control IP; AIP, miR-135a IP.