Figure 4.
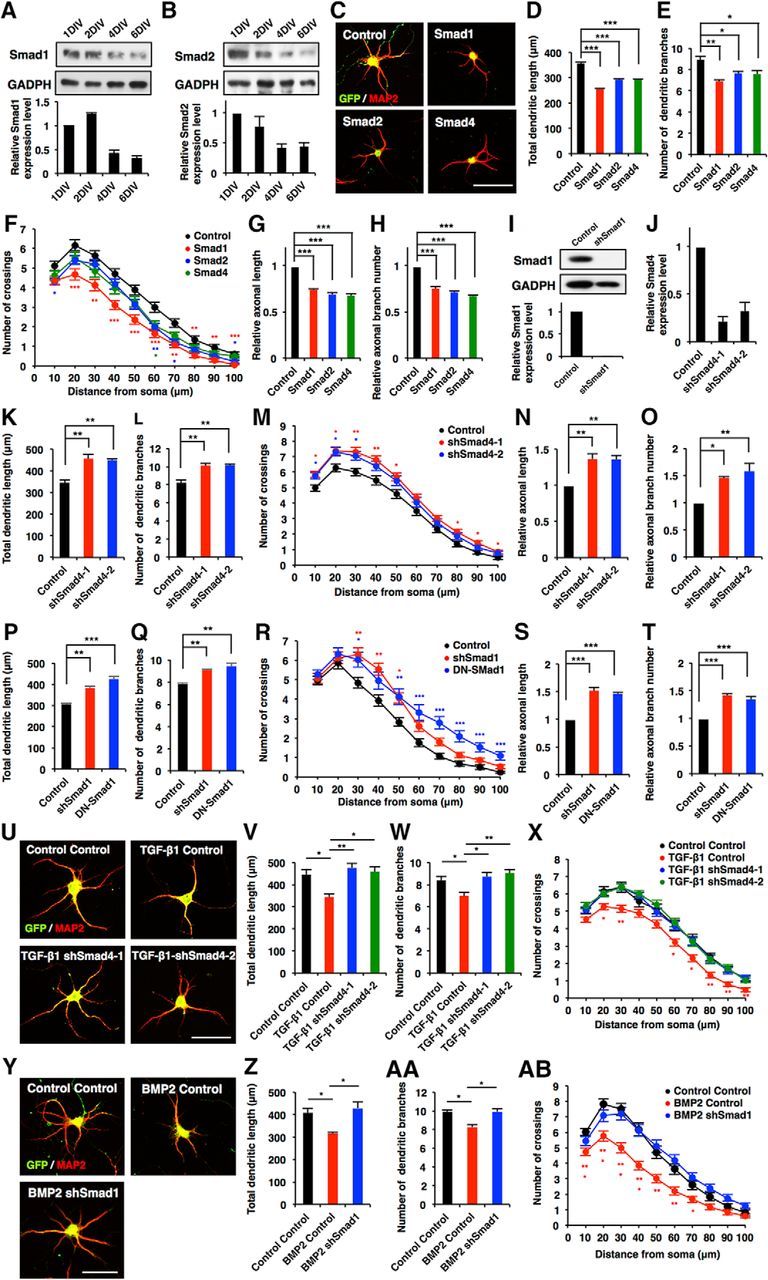
Canonical Smad-dependent signaling pathways suppress morphological development in hippocampal neurons. A, B, Western blot analysis of Smad1 and Smad2 expression in developing cultured hippocampal neurons. C, Representative images of GFP (green) and MAP2 (red) staining of 6DIV hippocampal neurons infected with lentiviruses expressing Smad1, Smad2, or Smad4. Scale bar, 50 μm. D, E, Quantification of total dendritic length (D) and branch numbers (E) of 6DIV hippocampal neurons infected with lentiviruses expressing Smad1, Smad2, or Smad4. F, Quantification of dendrite complexity by Sholl analysis of 6DIV hippocampal neurons infected with lentiviruses expressing Smad1, Smad2, or Smad4. G, H, Quantification of total axon length (G) and axon branch numbers (H) of 3DIV hippocampal neurons infected with lentiviruses expressing Smad1, Smad2, or Smad4. I, Hippocampal neurons were infected with lentiviruses expressing shRNA against Smad1. The expression of Smad1 protein was measured by Western blotting using an anti-Smad1 antibody. J, Validation of the effect of Smad4 shRNAs. qRT-PCR analysis of Smad4 levels in cultured hippocampal neurons infected with lentiviruses expressing shSmad4-1 or shSmad4-2. K, L, Quantification of total dendritic length (K) and branch numbers (L) of 6DIV hippocampal neurons infected with lentiviruses expressing shSmad4-1 and shSmad4-2. M, Quantification of dendrite complexity by Sholl analysis of 6DIV hippocampal neurons infected with lentiviruses expressing shSmad4-1 and shSmad4-2. N, O, Quantification of total axon length (N) and axon branch numbers (O) of 3DIV hippocampal neurons infected with lentiviruses expressing shSmad4-1 and shSmad4-2. P, Q, Hippocampal neurons were infected with lentiviruses expressing shSmad1 and DN-Smad1: quantification of total dendritic length (P) and branch numbers (Q). R, Quantification of dendrite complexity by Sholl analysis of 6DIV hippocampal neurons infected with lentiviruses expressing shSmad1 and DN-Smad1. S, T, Quantification of total axon length (S) and axon branch number (T) of 3DIV hippocampal neurons infected with lentiviruses expressing shSmad1 and DN-Smad1. U, Representative images of GFP and MAP2 staining of 6DIV hippocampal neurons treated with TGF-β1, and infected with lentiviruses expressing control shRNA, shSmad4-1 or shSmad4-2. Scale bar, 50 μm. V, W, Quantification of total dendritic length (V) and branch numbers (W) of 6DIV hippocampal neurons treated with TGF-β1, and infected with lentiviruses expressing control or shSmad4-1 and shSmad4-2. X, Quantification of dendrite complexity by Sholl analysis of 6DIV hippocampal neurons treated with TGF-β1, and infected with lentiviruses expressing control or shSmad4-1 and shSmad4-2. Y, Representative images of GFP (green) and MAP2 (red) staining of 6DIV hippocampal neurons treated with BMP2, and infected with lentiviruses expressing shSmad1. Scale bar, 50 μm. Z, AA, Quantification of total dendritic length (Z) and branch numbers (AA) of 6DIV hippocampal neurons treated with BMP2, and infected with lentiviruses expressing control or shSmad1. AB, Quantification of dendrite complexity by Sholl analysis of 6DIV hippocampal neurons treated with BMP2, and infected with lentiviruses expressing control or shSmad1. Data are presented as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 by one-way ANOVA, Tukey's post-test. N = 3 independent experiments; at least 50 neurons were analyzed in each experiment.
