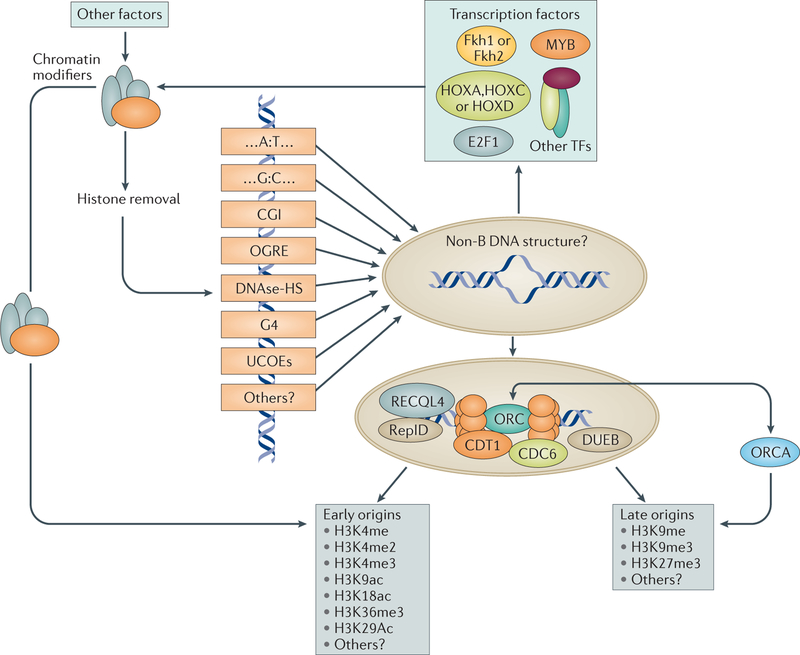
Figure 2. Genetic and epigenetic features of replication origins.
Although there are no strong consensus DNA sequences to anchor origins in metazoans, whole-genome sequence analyses have revealed several motifs that are present near or at origin sites. These include A:T- and G:C-rich motifs, CpG islands (CGIs), origin G-rich repeated elements (OGREs), DNase 1-hypersensitive sites (DNase-HS), quadruplex-like structures (G4) and ubiquitous chromatin-opening elements (UCOEs), which might increase the propensity of origins to unwind and adopt a non-B DNA structure. Groups of replication origins interact with proteins from the pre-replication complex and with proteins located at a subset of replication origins (such as replication-initiation determinant (RepID), DNA-unwinding element-binding protein B (DUEB) and RECQ-like DNA helicase type 4 (RECQL4)). Such interactions may also facilitate binding of transcription factors (TFs) such as forkhead box protein 1 (Fkh1), Fkh2, MYB, homeobox protein A (HOXA), HOXC, HOXD and E2F1) or other proteins that may regulate chromatin structure locally. Origins in early-replicating, open chromatin are preferentially enriched with euchromatin markers (such as H3K4 monomethylation (H3K4me), H3K4 dimethyation (H3K4me2), H3K4 trimethylation (H3K4me3), H3K9 acetylation (H3K9ac), H3K18ac, H3K36me3 and H3K29ac). Cell type-specific, late-replicating origins often associate with heterochromatin markers (such as H3K9me3, H3K27me3 and ORC-associated protein (ORCA), an ORC-binding protein that interacts with H3K9me3).