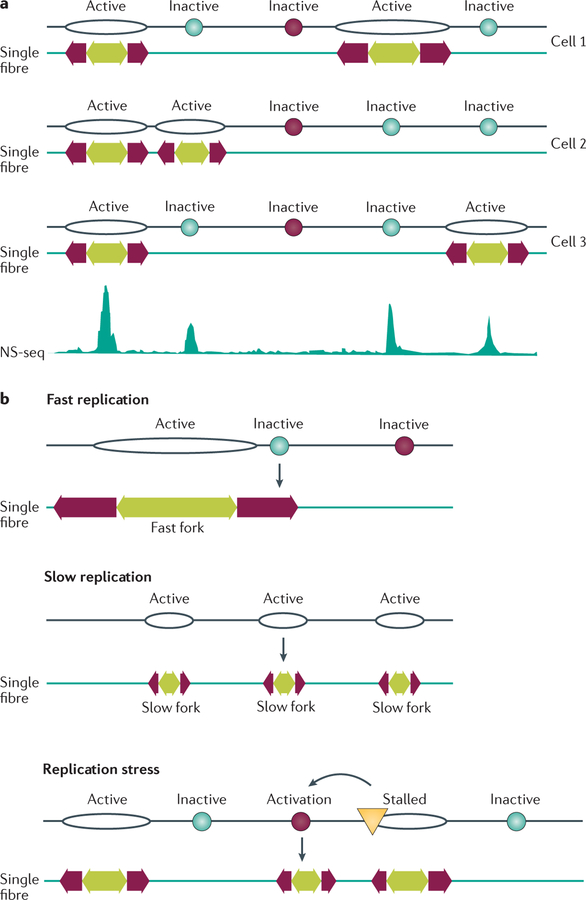
Figure 4. Replication origin choice and origin dormancy.
Constitutive origins (depicted as black oval-shaped outlines) initiate replication at all times in all cells; flexible origins (turquoise circles) initiate replication intermittently with initiation frequencies that vary from cell to cell; and dormant origins (dark red circles) are licensed to initiate replication but never, or very rarely, initiate during an unperturbed cell cycle. Dormant origins can be activated, however, if they reside next to a stalled or damaged replication fork (yellow triangle). a | Top: initiation from both constitutive and flexible origins shown on three hypothetical fibres, with each cell exhibiting a unique pattern of initiation. Experimentally, single-fibre analyses indicate the selection of initiation sites from the pool of flexible origins. Bottom: a population-based assay based on the abundance of nascent strands (nascent strand sequencing (NS-seq)) captures short, newly replicated DNA from all origins. Overall, the mean distances between population-based origin peaks are shorter than those measured on single fibres, indicating origin choice. b | Initiation rates can increase when DNA synthesis slows or when replication forks stall owing to DNA damage. Fast replication is associated with few origins on average because some potential initiation sites are passively replicated by adjacent replication forks before their own replication is actively initiated. Slow replication can increase the frequency of initiation from flexible origins, resulting in shorter inter-origin distances detected on single fibres. In addition, perturbed replication can activate dormant origins that do not normally initiate replication during unperturbed DNA synthesis.