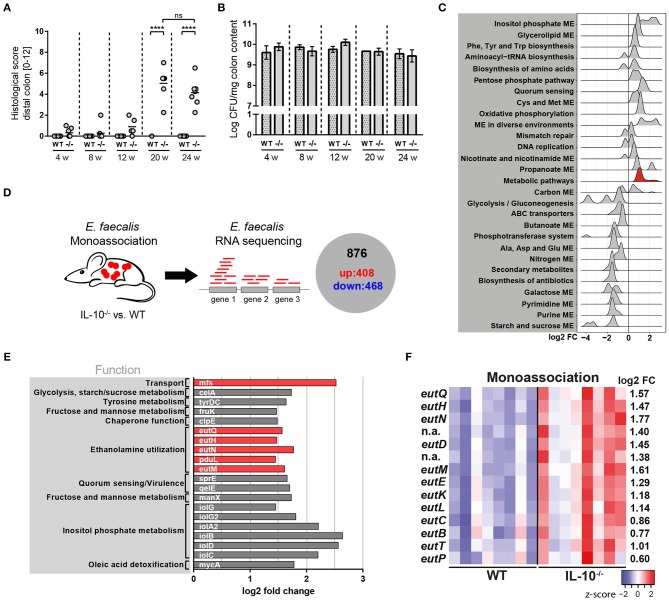
Figure 1.
E. faecalis adapts to a chronic inflammatory environment by altering its gene expression profile. (A,B) Wild type (WT) and IL-10−/− (–/–) mice were monoassociated with E. faecalis for 4, 8, 12, and 24 weeks (w). (A) Histological inflammation score in the distal colon. (B) E. faecalis counts/mg in luminal contents from colon. (C) Experimental setup: Transcriptional profiling of luminal bacteria isolated from the colon of E. faecalis-monoassociated IL-10−/− (inflamed) and healthy wild type mice (n = 8/group) revealed 408 significantly up-regulated genes and 486 down-regulated genes in E. faecalis under inflammatory conditions. (D–F) Expression profiles of E. faecalis in an inflamed (IL-10−/−) vs. a non-inflamed (WT) environment in monoassociated mice. (D) Expression distribution (log2 lower or higher) of genes for GSEA-enriched categories (KEGG). The gene group containing eut-genes is highlighted red. (E) Top 20 up-regulated genes. Only annotated genes with known function are shown. Genes of the eut locus are highlighted in red. Gene functions were assigned according to KEGG database. (F) Differential expression of genes in relation to a chronically inflamed environment is shown for significantly regulated eut genes. The log2 ratio of mean abundance (IL-10−/− vs. WT) of normalized expression levels is shown (up regulation is indicated red, down regulation is indicated blue). Differences were considered significant for ****p ≤ 0.0001.