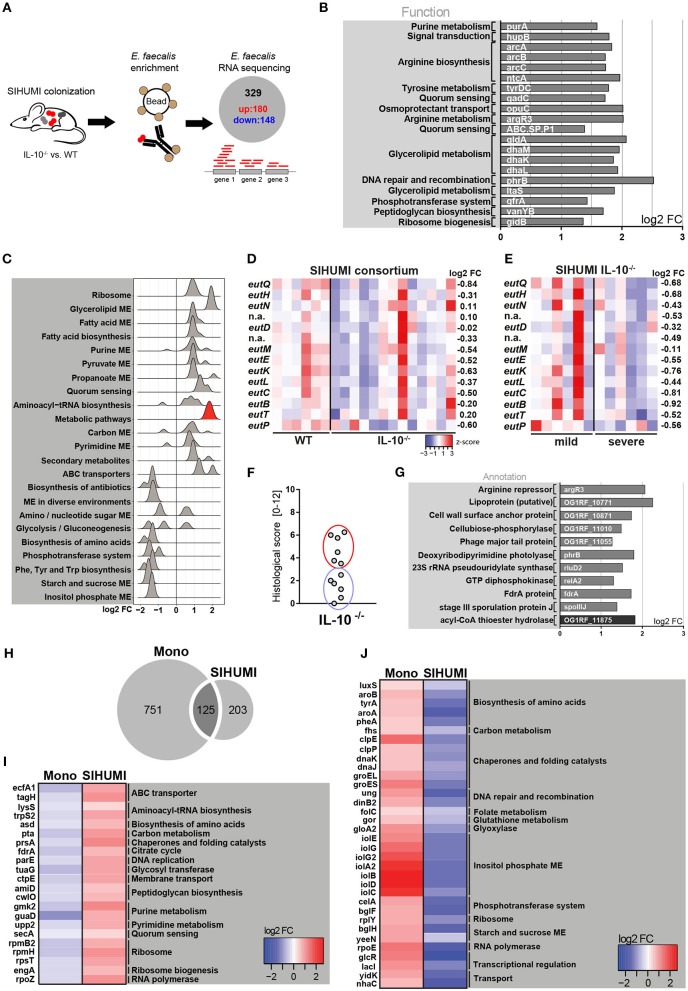
Figure 3.
Complex bacterial consortia reprogram E. faecalis gene expression in response to inflammation. Luminal E. faecalis cells were isolated from the colon of inflamed IL-10−/− (n = 13) and healthy wild type (WT) (n = 6) mice, colonized with SIHUMI consortium (16 weeks), using immuno-magnetic separation and subjected to RNA sequencing analysis. (A) Experimental setup: Transcriptional profiling revealed 180 significantly up-regulated and 148 significantly down-regulated genes in E. faecalis isolated from IL-10−/− mice compared to E. faecalis isolated from wild type mice colonized with the SIHUMI consortium. (B) Top 20 up-regulated genes in E. faecalis isolated from SIHUMI colonized mice (IL-10−/− vs. WT). Only annotated genes with known function are shown. Gene functions were assigned according to KEGG database. (C) Expression distribution of enriched genes for GSEA enriched categories (KEGG) in E. faecalis isolated from SIHUMI colonized mice (IL-10−/− vs. WT). The gene group highlighted red contains the eut-genes. (D) Differential expression of eut genes in E. faecalis of an inflamed vs. non-inflamed environment. The log2 ratios of mean abundance (IL-10−/− vs. WT) of normalized expression levels are shown (up regulation is indicated red, down regulation is indicated blue). (E) Differential expression of eut genes in E. faecalis of a severely inflamed vs. mildly inflamed environment. The log2 ratios of mean abundances of normalized expression levels are shown (up regulation is indicated red, down regulation is indicated blue). (F) Histological inflammation scores in the distal colon of SIHUMI colonized IL-10−/− mice comparing severely inflamed mice (red) and mildly inflamed mice (blue). (G) Significantly differentially regulated genes of E. faecalis isolated either from severely inflamed or mildly inflamed SIHUMI colonized IL-10−/− mice (up regulation is indicated gray, down regulation is indicated black). (H–J) Differentially regulated genes in response to inflammation of E. faecalis isolated from monoassociated (IL-10−/− vs. WT) compared to SIHUMI-colonized mice (IL-10−/− vs. WT). (H) Venn diagram showing the differentially regulated genes shared. (I) Genes down regulated in monoassociated and up regulated in SIHUMI-associated mice, respectively. The log2 ratios of mean abundances of normalized expression levels are shown (up regulation is indicated red, down regulation is indicated blue). Gene functions were assigned according to the KEGG database; only annotated genes with known function are shown. (J) As before, but genes up regulated in monoassociated and down regulated in SIHUMI associated mice, respectively.