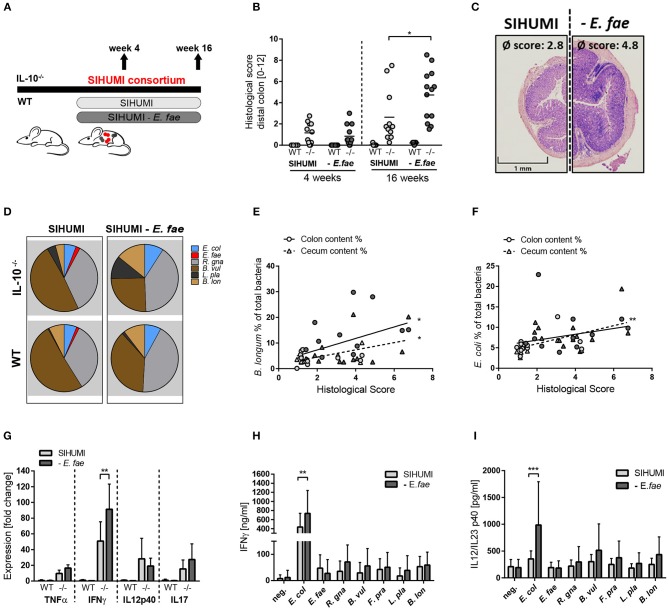
Figure 4.
Complex microbial consortia interactions reprogram colitogenic activity of E. faecalis in gnotobiotic IL-10−/− mice. (A) Experimental setup: Germ-free IL-10−/− (–/–) and wild type (WT) mice were colonized with SIHUMI consortium or SIHUMI consortium without E. faecalis (- E. fae) for a period of 4 and 16 weeks. (B) Histological inflammation in the distal colon. (C) Representative hematoxylin/eosin-stained sections of distal colon after 16 weeks of colonization. (D) Relative abundances of SIHUMI bacterial species in luminal colon content by 16S rRNA gene targeted qPCR. (E) Correlation between B. longum abundance (colon and cecum luminal content) and mean histological scores of cecum tip and distal colon. (F) As before, correlation of E. coli abundance. (G) Cytokine expression in colon tissue sections shown as fold change, normalized to cytokine expression levels in wild type mice colonized with the SIHUMI consortium. (H) IFNγ secretion of MLN cells isolated from SIHUMI or SIHUMI without E. faecalis colonized mice (colonization period: 16 weeks) that were re-activated with the respective bacterial lysate for 72 h. (I) As before, IL-12p40 secretion. E. col, Escherichia coli; E. fae, Enterococcus faecalis; R. gna, Ruminococcus gnavus; B. vul, Bacteroides vulgatus; L. pla, Lactobacillus plantarum; B. lon, Bifidobacterium longum. Differences were considered significant for *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.