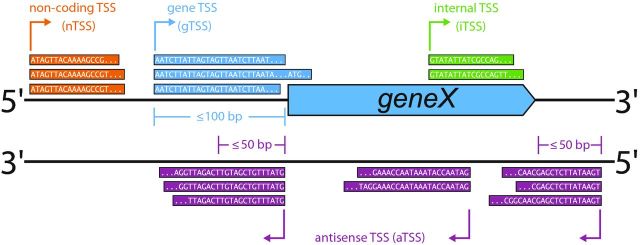
Figure 1.
TSS classification based on fixed-length thresholds. Based on fixed-length thresholds, a TSS can be annotated as gTSS, aTSS, iTSS or nTSS. In this example, a TSS is classified as gTSS (gene TSS, TSS and exemplary sequencing reads are shown in blue) if it is at most 100 bp upstream of a protein-coding gene. An aTSS (antisense TSS, shown in purple) must be within 50 bp of a protein coding gene or directly antisense to it, and an iTSS (internal TSS, shown in green) must be located in sense orientation within a coding sequence. A TSS that is located in IGRs and thereby does not match any of the previous criteria is classified as nTSS (non-coding TSS, shown in orange) that gives rise to sRNA candidates (another common nomenclature for this class is orphan TSS or oTSS; Sharma et al.2010; Dugar et al.2013).