Figure 3.
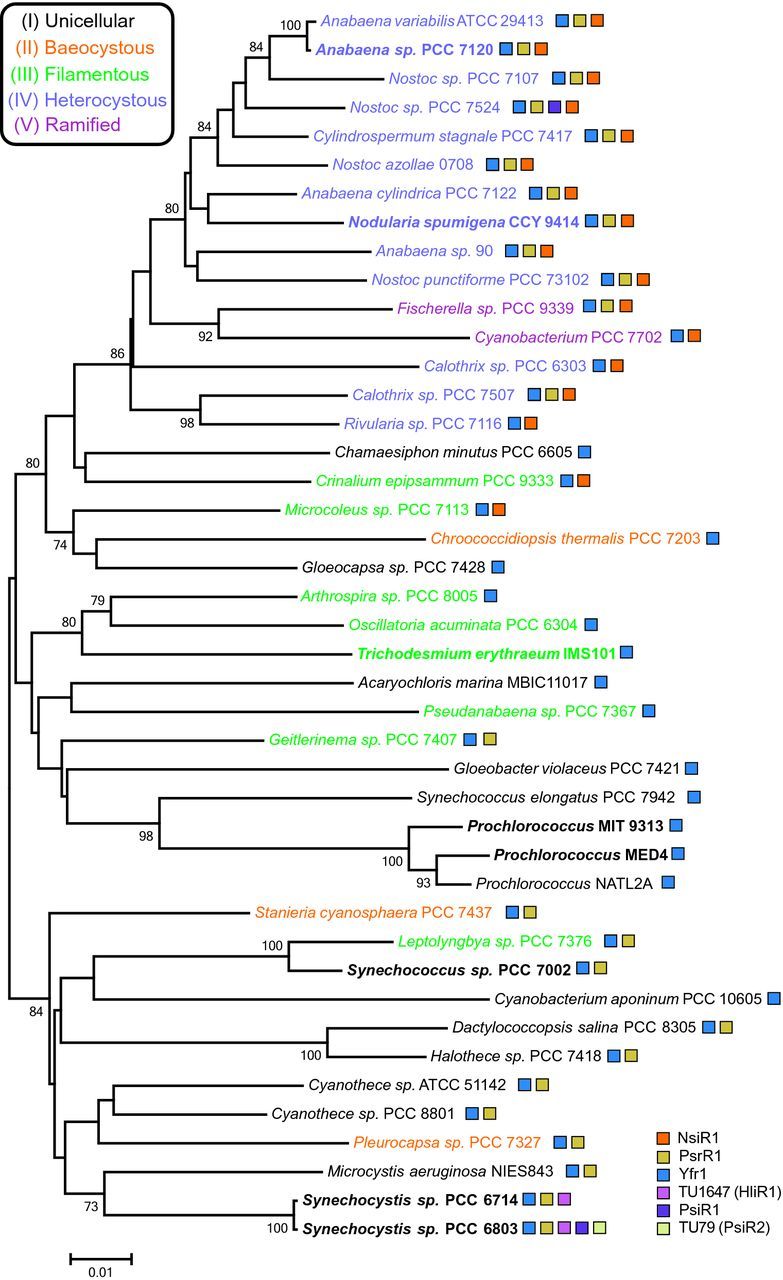
Unrooted cyanobacterial species tree and the distribution of selected sRNAs. The tree is based on an alignment of 16S rRNA genes from selected cyanobacteria. Major morphological characteristics are color coded according to the designations by Shih et al. (2013). Bootstrap values ≥70% are given at the respective nodes. The distribution of four non-coding sRNAs and two putative coding small mRNAs (PsiR1 and HliR2) is indicated by colored rectangles. The non-coding sRNA NsiR1 was first described in Anabaena sp. PCC 7120 and verified to exist in at least 19 other cyanobacteria that share the capability for nitrogen fixation and heterocyst differentiation (Ionescu et al.2010). NsiR1 is transcribed from a tandem array of direct repeats upstream of hetF (Ionescu et al.2010), a known regulator of heterocyst differentiation (Wong and Meeks 2001). NsiR1 expression is controlled by NtcA and HetR, two transcription factors critical for N2 fixation and heterocyst development and is restricted to developing heterocysts (Ionescu et al.2010; Muro-Pastor 2014). The non-coding sRNA Yfr1 (cyanobacterial functional RNA1) was initially identified in marine picocyanobacteria (Axmann et al.2005) and later found to be widely distributed throughout the cyanobacterial phylum (Voss et al.2007). Yfr1 has been suggested to control sbtA expression, which encodes the sodium-dependent bicarbonate transporter SbtA in Synechococcus elongatus PCC 6301 (Nakamura et al.2007) and outer membrane proteins (soms or porins) in Prochlorocococus sp. MED4 (Richter et al.2010). PsrR1 was initially predicted by comparative genome analysis (Georg et al.2009) and has recently been functionally characterized (Fig. 4 and (Georg et al.2014).
