Figure 7.
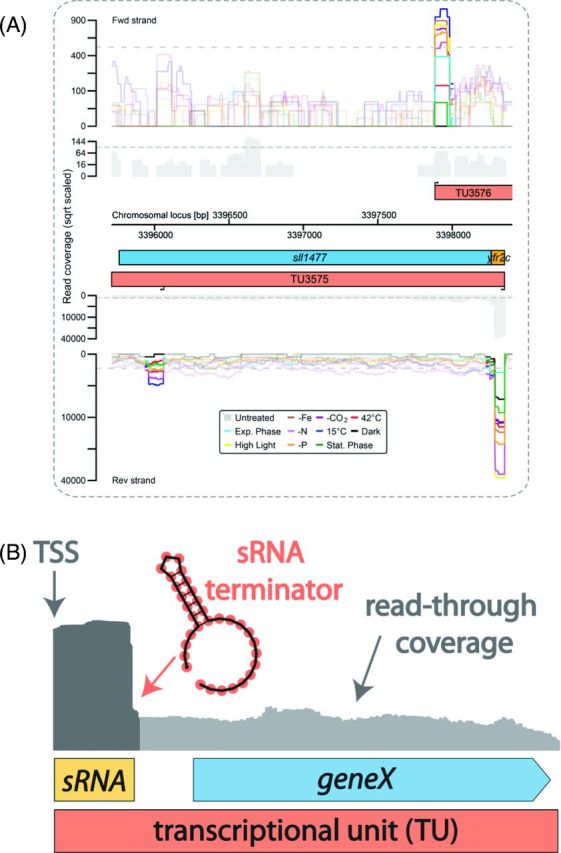
The actuaton concept. (A) The transcriptional organization of the yfr2c actuaton region in Synechocystis sp. PCC 6803. The colored graphs represent the accumulation of treated reads from a dRNA-seq analysis of Synechocystis sp. PCC 6803 under 10 different conditions (Kopf et al.2014a). All TSS positions inferred from the TEX-treated cDNA library are indicated by black arrows, and the graphs for the following 100 nt are highlighted. Transcriptional units (TUs, red) were inferred from the untreated read coverage (gray). Protein-coding genes are displayed in blue and previously predicted sRNAs are shown in yellow (Mitschke et al.2011b). Data for the forward strand is shown above and data for the reverse strand is shown below the axis that shows the chromosomal locus in bp. A gTSS upstream of sll1477 gives rise to both the sRNA Yfr2c (Voss et al.2009) and the mRNA for protein Sll1477, a protease of the abortive phage infection (abi, CAAX) family, and the respective TU3575 is therefore classified as an actuaton. Further downstream, TU3575 contains an internal start site (iTSS). On the forward strand, the TU3576 TSS gives rise to a weakly expressed asRNA to TU3575. The arrangement is well conserved in the closely related strain Synechocystis sp. PCC 6714 (Kopf et al.2015a), with the exception of the exact TSS position of the asRNA, which is slightly different. (B) Model of the actuaton concept. The expression of a downstream gene is driven by read-through (RNA-seq coverage shown in light gray) over the terminator from an sRNA that accumulates as a discrete and abundant transcript (RNA-seq coverage shown in dark gray). Because the downstream gene lacks an own TSS, it is classified in a joint transcriptional unit (TU) together with the sRNA.
