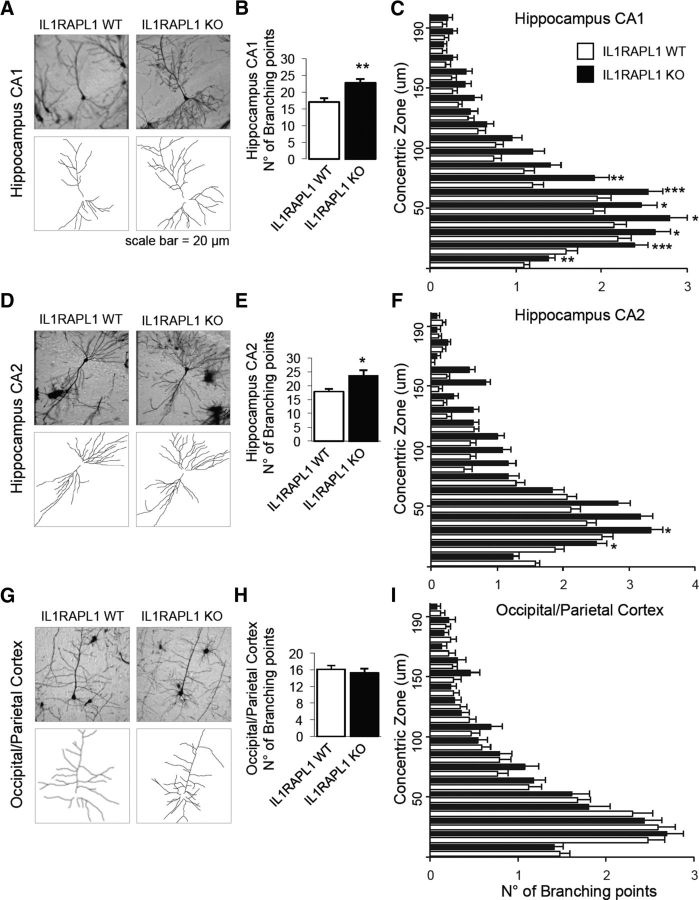
Figure 3.
Dendritic arborization complexity of hippocampal neurons is altered in Il1rapl1-KO adult mice. A, D, G, Representative images of Golgi-stained pyramidal neurons from WT and Il1rapl1-KO mice in CA1 (A), CA2 (D), and occipital/parietal cortex (G) and a schematization of their dendrites below. B, E, H, The graphs show the quantification of total number of neuronal branching points from WT and Il1rapl1-KO mice (8 neurons analyzed for each mouse; N = 4–9 for each genotype) from CA1 (B), CA2 (E), and occipital/parietal cortex (H). Data are shown as mean of number of branching points ± SEM. *p < 0.05, **p < 0.005 vs corresponding WT, Student's t test. C, F, I, Quantification of the number of branching points from WT and Il1rapl1-KO mice plotted on the distance from the soma (μm). All values represent mean of total branching points every 10 μm ± SEM. *p < 0.05, **p < 0.005, ***p < 0.001 vs corresponding WT, Student's t test.