Figure 5.
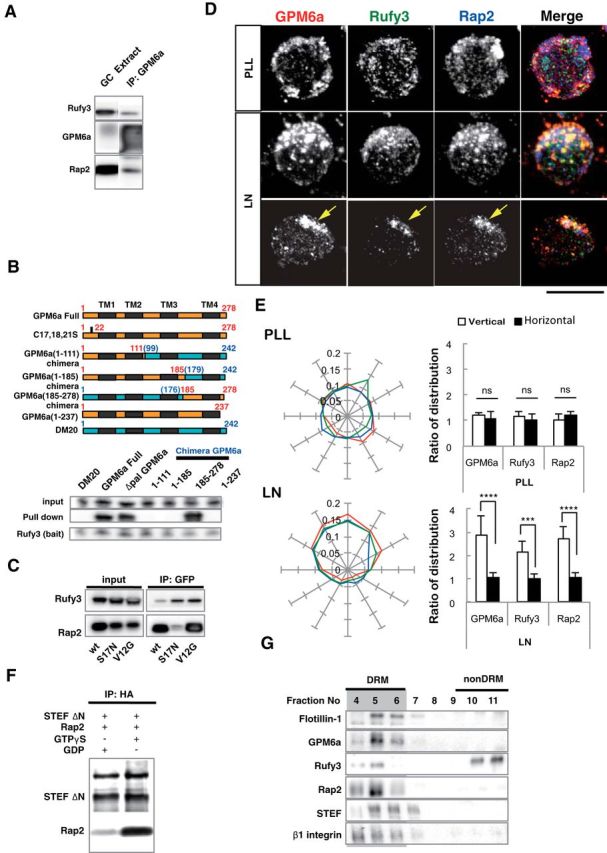
GPM6a-RUFY3-Rap2-STEF/Tiam2 forms a protein complex that is enriched in lipid rafts. A, Immunoprecipitation (IP) of the Triton X-100 extracts of growth cone (GC) fractions using an anti-GPM6a Ab. The blots were immunostained with Abs against GPM6a, Rufy3, and Rap2. The RUN domain protein Rufy3 and a small GTPase, Rap2, were coprecipitated with GPM6a, suggesting the presence of a GPM6a-Rufy3-Rap2 ternary complex in vivo. B, Identification of the Rufy3-binding region of GPM6a using GPM6a mutants. Top, To maintain the four-transmembrane domain in the mutants, chimeras of GPM6a and DM20, another member of the PLP family, were constructed. The fragments derived from GPM6a (orange) were joined with those from DM20 (blue). The amino acid numbers of GPM6a and DM20 are shown in red and blue respectively. Transmembrane domains are shown in black. (See Sato et al., 2011). Bottom, Constructs containing EGFP-tagged full-length GPM6a (aa 1–278), Δpal-GPM6a, chimeras, or a C-terminus deletion (aa 1–237) were coexpressed with myc-Rufy3 in COS-7 cells. Lysates were immunoprecipitated with an anti-myc antibody. The input was Western blotted with an anti-myc (Rufy3, bait) or an anti-GFP antibody (input) and the immunoprecipitate (pull-down) was Western blotted using an anti-EGFP (input) and myc Abs (bait). The Rufy3-binding site of GMP6a was determined as being located within GPM6a aa 185–278. C, Characterization of Rap2 binding to Rufy3. FLAG-tagged Rap2, the GDP-binding DN form (S17N), or the GTP-binding CA form (V12G) was preincubated with EGFP-Rufy3. EGFP-Rufy3 was immunoprecipitated with an anti-EGFP Ab and coprecipitated Rap2 was detected by Western blotting with anti-FLAG. The GDP-bound form of Rap2 lost the ability to bind Rufy3. D, GPM6a, Rufy3, and Rap2 are colocalized in cortical neurons grown on LN 30 minutes after plating of cortical neurons on LN but not on PLL. Iimmunofluorescence analysis indicated that the asymmetrically assembled endogenous GPM6a (red) was colocalized with endogenous Rufy3 (green) and Rap2 (blue). Arrows indicate their cellular accumulation sites. Scale bar, 20 μm. E, Left, Cumulative angular probability distribution of fluorescence of the GPM6a (red), Rufy3 (green), and Rap2 (blue) in Figure 3D. Right, Ratio of the fluorescence intensities of GPM6a, Rufy3, and Rap2 on the vertical or horizontal axis is shown. Asymmetric localization of each protein was specifically observed in neurons on LN. Data are shown as means ± SD (n = 20 for each; two-tailed t test, vertical vs horizontal; LN-GPM6a, 2.90 ± 0.79 vs 1.07 ± 0.20; LN-Rufy3, 2.17 ± 0.47 vs 1.03 ± 0.18; LN-Rap2, 2.70 ± 0.56 vs 1.06 ± 0.19). ***p < 0.001. F, GTP-bound Rap2 specifically interacts with the Rac1 GEF Tiam2/STEF. HA-STEF was preincubated with FLAG-Rap2 in the presence of GTPγ with FLAG. HA-STEF was immunoprecipitated using an anti-HA antibody and coprecipitated Rap2 was detected by blotting with an anti-FLAG Ab. G, The complex components downstream of GPM6a were distributed in the DRM. Mouse brains were lysed and subjected to a sucrose density gradient and collected fractions were Western blotted for the indicated proteins. The DRM marker flotillin 1 was detected in fractions #4 to #6; GPM6a, Rap2, STEF, and β1 integrin (a LN receptor) were enriched in DRMs and Rufy3 was also found in these fractions.
