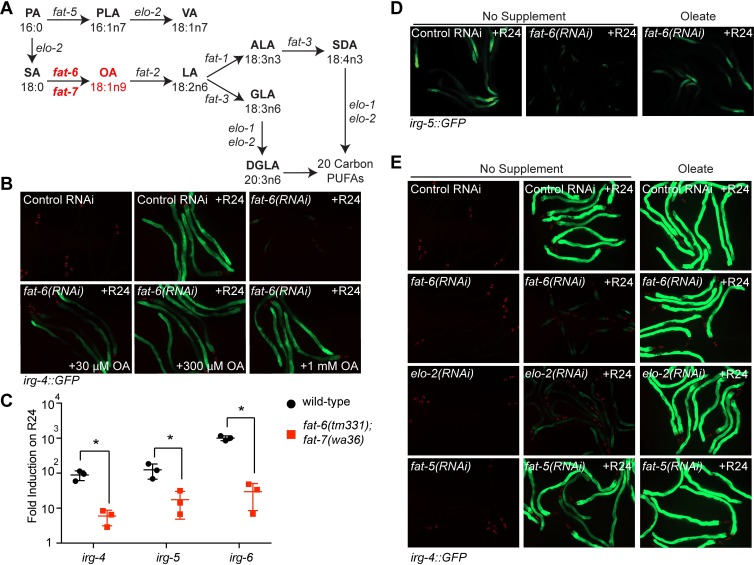
Fig 1. An RNAi screen identifies a role for MUFAs in the activation of C. elegans innate immune effectors.
A. A schematic of the long-chain fatty acid synthesis pathway in C. elegans adapted from [6,17]. Fatty acid nomenclature: X:YnZ, X indicates the number of carbon atoms, Y denotes the number of double bonds, and Z designates the position of the terminal double bond relative to the methyl end of the molecule. Abbreviations: ALA, α-linoleic acid; DGLA, di-homo-γ-linoleic acid; GLA, γ-linoleic acid; LA, linoleic acid; OA, oleate; PA, palmitic acid; PLA, palmitoleic acid; SA, stearic acid; SDA, stearidonic acid; VA, vaccenic acid. B. The C. elegans irg-4::GFP immune reporter was exposed to control or fat-6(RNAi) bacteria seeded on plates containing control or the indicated levels of oleate. The animals were then transferred at the L4 stage to plates containing R24 for approximately 18 hours. Red pharyngeal expression is the myo-2::mCherry co-injection marker, which confirms the presence of the transgene. C. Expression of the immune effectors irg-4, irg-5, and irg-6 were determined using qRT-PCR in wild-type or fat-6(tm331);fat-7(wa36) mutant animals exposed to R24 or solvent control (1% DMSO) for six hours. Data are the average of three independent replicates, each normalized to a control gene and presented as fold induction in R24-exposed vs. solvent control-exposed animals in the indicated genetic background. Error bars show standard deviation. Significance was determined by t-tests, * p<0.05. D. Animals containing the irg-5::GFP immune reporter were grown as described above and exposed to R24. All animals had a Rol phenotype, which confirms the presence of the transgene. E. C. elegans containing the irg-4::GFP immune reporter were fed RNAi bacteria targeting the indicated genes on plates containing control or oleate and exposed to R24, as described above.