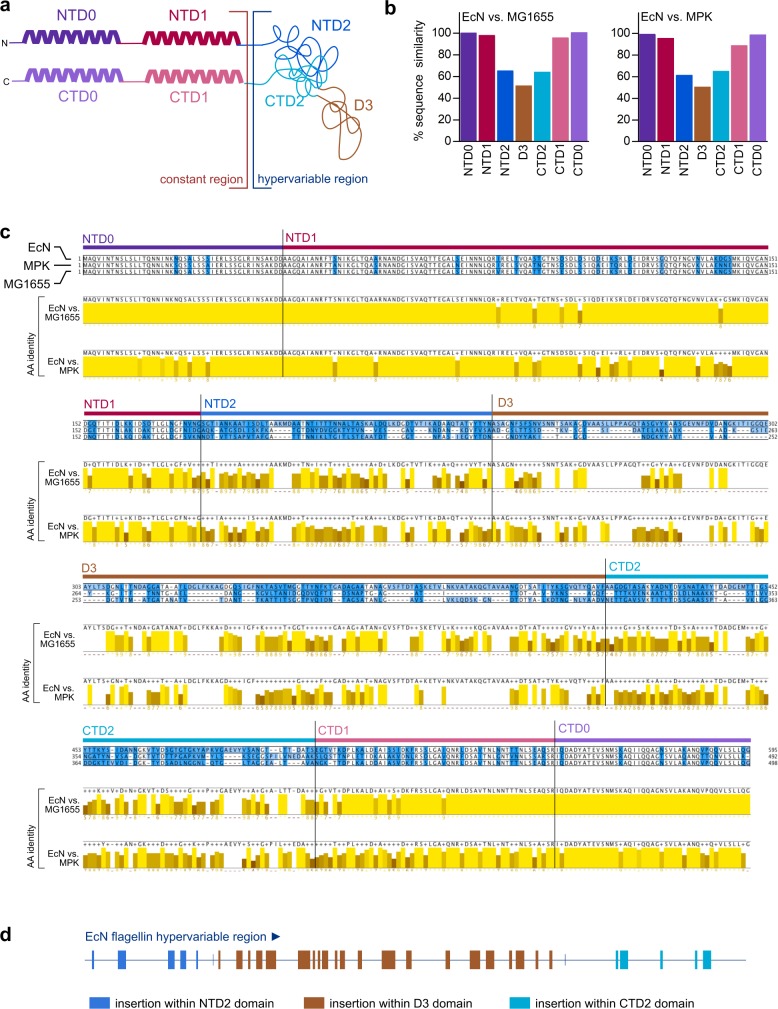
Fig 3. Detailed comparison of the flagellin amino-acid sequences of different E. coli strains.
Protein alignment of FliC proteins from EcN (CCQ05465.1), MPK, and MG1655 (NP_416433.1) genomes. (a) Schematic structure of flagellin according to Yonekura and colleagues [50,51]. D0 and D1 comprise conserved N- and C-termini of fliC, packed into α-helical structures in the filament core. The NTD2/CTD2- and D3-domain–containing HVR is attached adjacent to the D2 domains, located at the outer surface of the filament. (b) Quantification of amino-acid sequence similarities of all 6 flagellin domains. Computation was performed using similarity indices depicted in 3c. (c) The alignment was generated using MAFFT. Amino acids were colored by overall conservation (white: fully conserved, light blue to dark blue: high to low conservation). Consensus sequence and sequence conservation for pairwise comparisons of EcN to MPK and MG1655, respectively, are shown in yellow. Darker shades of yellow represent lower sequence conservation. (d) Schematic overview of the differences of the EcN flagellin HVR compared to the HVRs of both MG1655 and MPK. Sequences (insertions) that are only present in EcN HVR and not in MPK HVR or MG1655 HVR are highlighted as colored squares. AA, amino acid; CTD, C-terminal domain; EcN, E. coli Nissle 1917; fliC, flagellin; HVR, hypervariable region; MAFFT, Multiple Alignment using Fast Fourier Transform; MG1655, E. coli K12 MG1655; MPK, E. coli mpk; NTD, N-terminal domain.