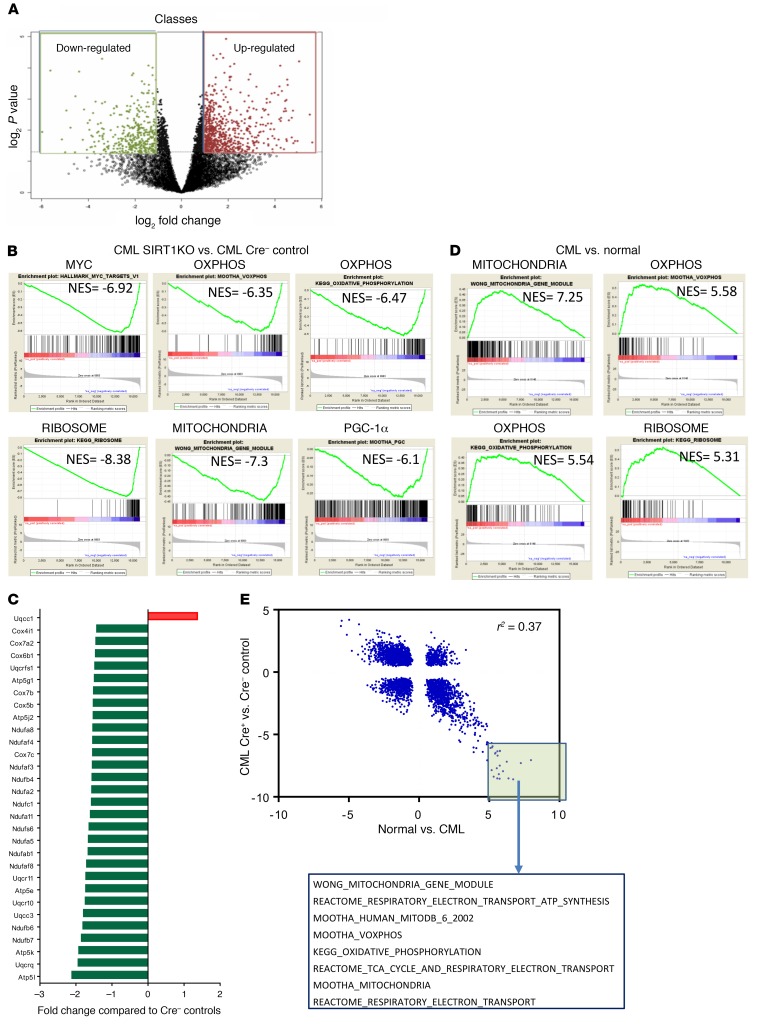
Figure 6. SIRT1 deletion inhibits mitochondrial gene expression in CML and normal hematopoiesis.
(A) Volcano plot showing differentially expressed genes in SIRT1-deleted (SIRT1 KO) compared with control (WT) CML stem cells (LSK) on RNA-Seq analysis. 1,063 Differentially expressed genes (2× fold change and P < 0.05) are highlighted. (B) GSEA analysis of gene expression data showing significant negative enrichment (downregulation) of Myc targets, ribosomal, oxidative phosphorylation, mitochondria, and PGC-1α–related gene sets in SIRT1-KO compared with control CML stem cells. (C) Downregulation of mitochondrial electron transport chain complex genes in SIRT1-deleted compared with control CML stem cells (red signifies upregulation, and green signifies downregulation). (D) GSEA analysis of gene expression data showing significant positive enrichment (upregulation) of mitochondrial, oxidative phosphorylation, and ribosomal gene sets in CML compared with normal stem cells. (E) Pearson’s correlation of NES for the 3567 GSEA signatures derived from analysis of normal versus CML compared with SIRT1-KO versus WT stem cells. The highlighted box indicates gene sets that are most highly enriched in the CML LTHSC versus normal LTHSC analysis and negatively enriched in the SIRT1 WT versus SIRT1-KO LSK analysis, and the mitochondria-related gene sets present among these, are shown.