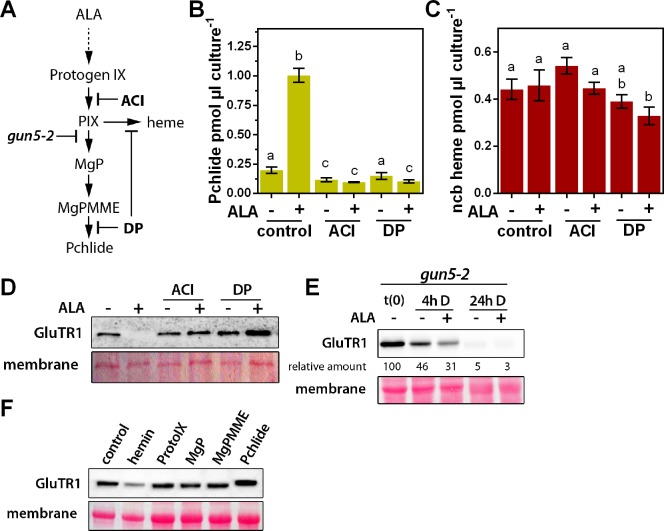
Figure 4. Identification of the stimulus responsible for triggering GluTR1 degradation.
A photoautotrophic Arabidopsis cell culture was used, together with mutants for the tetrapyrrole biosynthesis pathway, and isolated chloroplasts. (A) Overview of the tetrapyrrole biosynthesis pathway. Acifluorfen (ACI) prevents the formation of protoporphyrin IX (PIX) by inhibiting protoporphyrinogen oxidase. The iron chelator 2,2'-dipyridyl (DP) inhibits the reactions catalyzed by the ferrochelatases (FC) and aerobic cyclase, respectively. Knockout of GUN5 (gun5-2) prevents synthesis of Mg-protoporphyrin IX (MgP) and downstream intermediates. (B, C) Pchlide (B) and ncb heme (C) contents of photoautotrophically cultured Arabidopsis cells (PA) incubated in the presence (+) or absence of (-) 1 mM ALA. In addition to ALA, cells were treated with 100 µM ACI or 500 µM DP, respectively. PA cultures were pre-incubated with ACI or DP for 2 hr in darkness, before the buffer was exchanged for media supplemented with ACI, DP, and ALA. Treatment was performed for 16 hr in the dark. Data are given as mean ± sd (n = 4). Letters indicate statistical groups determined by Student’s t-test (p<0.05). (D) Levels of GluTR1 in cell cultures incubated in the absence (- ALA) or presence (+ALA) of added ALA and with or without ACI or DP, respectively. (E) Levels of GluTR1 in leaves of gun5-2 knockout mutants incubated without (-) or with (+) 1 mM ALA in the dark (D) for the indicated times. Signal intensities for GluTR1 relative to t(0) are shown. (F) Arabidopsis Col-0 chloroplasts were isolated and incubated for 6 hr in darkness in the presence of hemin and intermediates of the tetrapyrrole biosynthesis pathway. Pchlide was used at 1 µM and heme and the other porphyrins at 5 µM final concentration.
Figure 4—figure supplement 1. Phenotype (A) and genotyping (B) of Col-0 and the gun5-2 mutant.