Table 5.
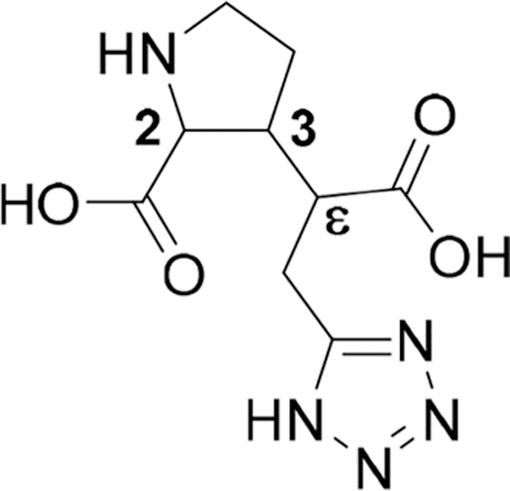
Binding Affinities of 3q-s1−4 at Native NMDA Receptors and Functional Characterization as Antagonists at Cloned GluN1/GluN2A-D Receptorsa
 | |||||||
|---|---|---|---|---|---|---|---|
|
Ki |
|||||||
| compd | 2,3-trans configuration | Abs stereochem or d.r. at ε-carbon | NMDA (binding affinity) | GluN1/GluN2A | GluN1/GluN2B | GluN1/GluN2C | GluN1/GluN2D |
| 3q-s1 | (−)-Abs (n.d.) | 96:4 | 28 [4.56 ± 0.06] | − | − | − | − |
| 3q-s2 | (−)-Abs (n.d.) | 4:96 | 1.3 [5.89 ± 0.09] | 1.2 ± 0.1 | 2.8 ± 0.1 | 6.5 ± 0.6 | 15 ± 2 |
| 3q-s3,4 | (+)-Abs (n.d.) | 1:2 | 0.25 [6.61 ± 0.07] | 0.61 ± 0.02 | 2.7 ± 0.1 | 12 ± 1 | 62 ± 9 |
All values in μM. Binding data are mean values of at least three individual experiments performed in triplicate. For NMDA: Ki values with mean pKi ± SEM in brackets. For GluN1/GluN2A-D, Ki values are reported as the mean ± SEM from six oocytes and were estimated from IC50 values using the Cheng−Prusoff relationship (see Methods). −: not tested; n.d.: not determined.
