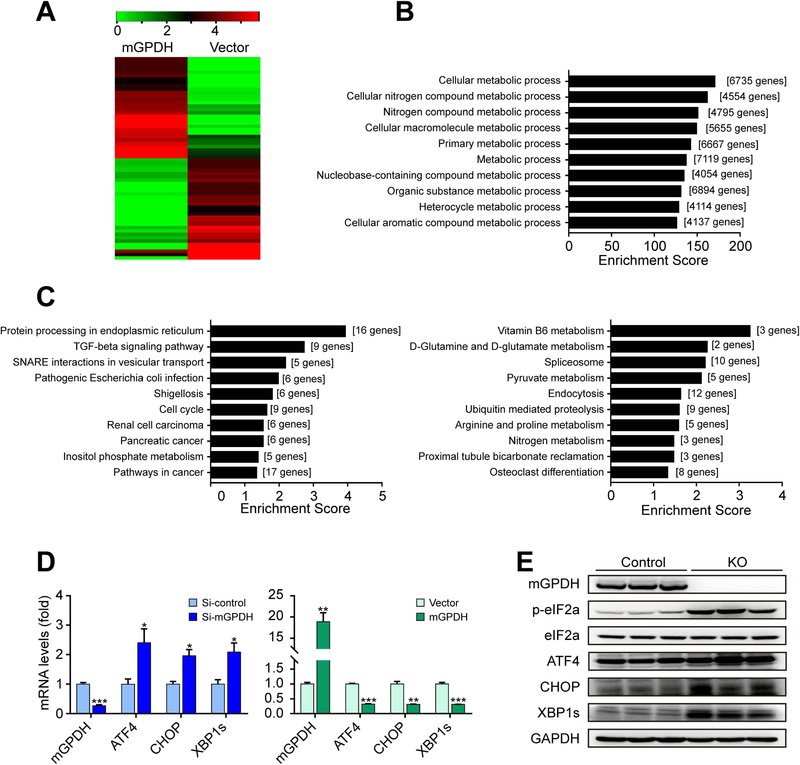
Figure 4. mGPDH modulates ER-related proteins.
(A-C) LO2 cells were transfected with pCMV3-N-GFPSpark-mGPDH plasmid or vector, and then treated with FFA for 24 h. The heatmap displays the top 30 up- and down-regulated genes (A). GO analysis was conducted with all altered genes, and the ten highest ranking biological process (BP) terms were shown (B). The ten highest ranking terms from KEGG pathway analyses of all significantly down-regulated (left) and up-regulated (right) (change-fold over 1.5) genes were shown (C). (D) LO2 cells were transfected with siRNA or plasmid for mGPDH inhibition or overexpression and then treated with FFA for 24 h, the mRNA levels of mGPDH and ER stress markers (ATF4, CHOP and XBP1s) were detected by qRT-PCR. (E) The hepatic expression of mGPDH and ER stress markers (p-eIF2α, ATF4, CHOP and XBP1s) in HFD-fed KO and control mice was detected by western blot. n = 3 for A-D, n = 6 mice per group for E. Data are presented as the mean ± S.E.M. *P < 0.05, **P < 0.01, ***P < 0.001.