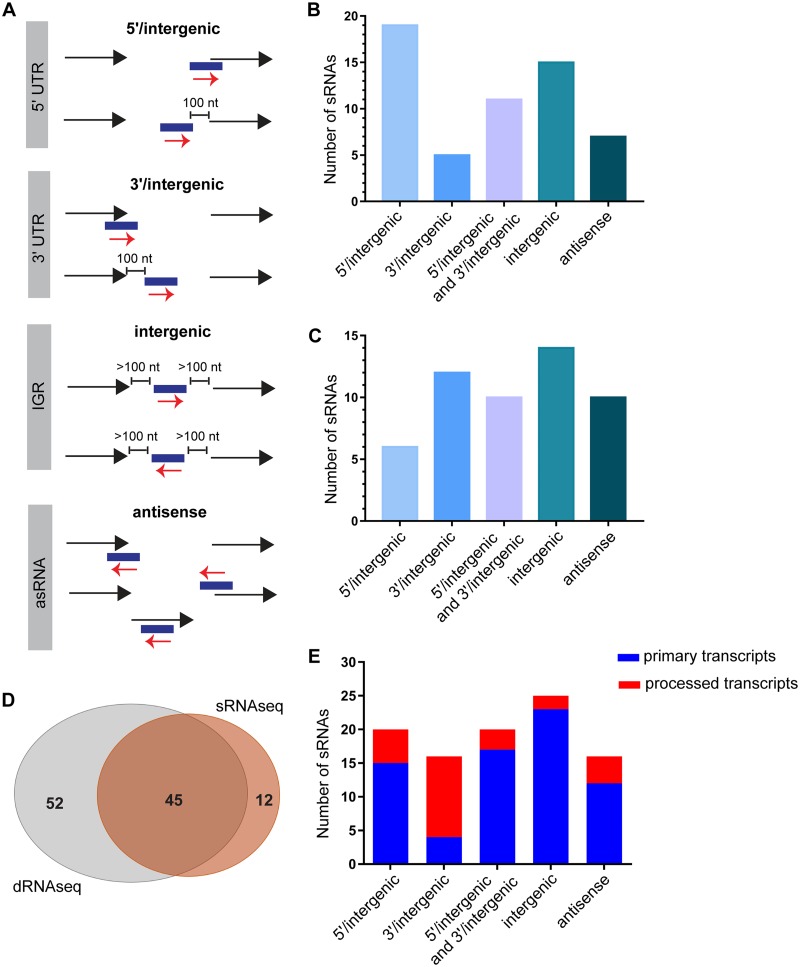
FIG 1.
Overview of the sRNAs identified in D39W using sRNA-seq and dRNA-seq. (A) Genomic context of the sRNAs identified in D39W. sRNAs identified in this study were classified into four major classes based on their location in the D39W genome relative to flanking genes. sRNAs partially overlapping the 5′ end of an annotated gene or located within a 100-nt distance of the start codon of the downstream ORF were classified as 5′/intergenic. sRNAs partially overlapping the 3′ end of an annotated gene or within a 100-nt distance from the stop codon of the upstream ORF were classified as 3′/intergenic. sRNAs were classified as intergenic if they were located at least 100 nt away from the flanking genes. sRNAs encoded on the opposite strand of an ORF and overlapping by at least 1 nt were classified as antisense sRNAs. IGR, intergenic region. (B) Genomic contexts of sRNAs identified by sRNA-seq analysis as defined above for panel A. (C) Genomic contexts of sRNAs identified by dRNA-seq analysis, but not sRNA-seq analysis, as defined above for panel A. (D) Venn diagram showing the overlap between the sRNAs identified by sRNA-seq and those identified by dRNA-seq analyses. (E) Transcription start site (TSS) characteristics of the sRNAs in each category defined above for panel A that were identified by dRNA-seq in D39W. sRNAs determined by dRNA-seq analysis to have unprocessed 5′ ends or to contain processed 5′ ends are indicated in blue and red, respectively.