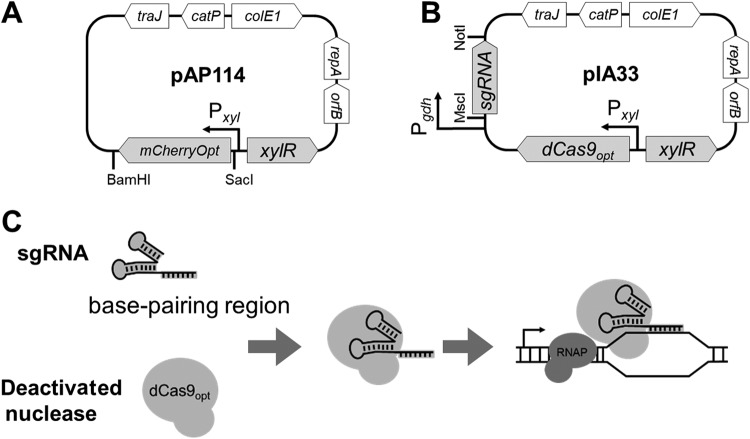
FIG 1.
Genetic maps and CRISPRi principle. (A) pAP114 for xylose-inducible expression. The expression of the red fluorescent protein mCherryOpt is under Pxyl control. The gene can be replaced using the SacI and BamHI restriction sites. (B) pIA33 for CRISPRi. Single guide RNA (sgRNA) is under Pgdh control, and dCas9 is under Pxyl control. The sgRNA sequence can be replaced using the MscI and NotI restriction sites. (C) CRISPRi principle. The deactivated nuclease binds sgRNA and is targeted to the gene of interest, but it is unable to cleave the DNA. Instead, dCas9 serves as a transcriptional roadblock. Features depicted are as follows: mCherryOpt encodes a red fluorescent protein variant that is codon optimized for expression in low-GC-content bacteria; dCas9 encodes the deactivated CRISPR nuclease codon optimized for expression in low-GC-content bacteria; orfB and repA are the replication regions; colE1 is the replication region of the E. coli plasmid pBR322 modified for a higher copy number; catP is the chloramphenicol acetyltransferase gene from Clostridium perfringens, conferring resistance to thiamphenicol in C. difficile or chloramphenicol in E. coli; and traJ encodes a conjugation transfer protein from plasmid RP4 (23). RNAP, RNA polymerase.