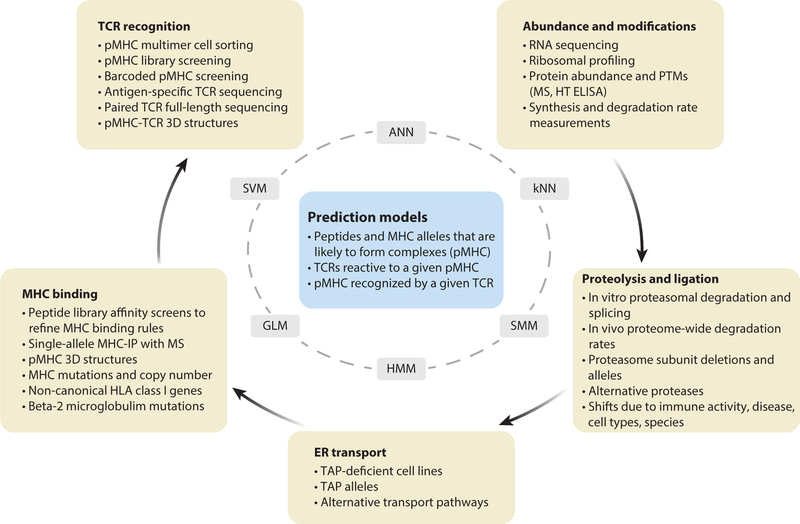
Figure 2.
Experimental tools and datasets needed to develop predictive models of antigen presentation and TCR recognition. The boxes in the outer circle represent steps in the process of MHC-I antigen presentation and recognition, and the experimental strategies and types of datasets that need to be collected to train predictive models. The inner circle lists some of the analytical methods for learning the underlying rules that govern each step or the integrated process. The box in the center shows the output (or goals) of the predictive models. Abbreviations: ANN, artificial neural networks; GLM, generalized linear models; HMM, hidden Markov models; HT, high throughput; kNN, k-nearest neighbors; MHC-IP, immunoprecipitation of MHC proteins from cells; MS, mass spectrometry; pMHC, peptide-MHC complex; PTMs, posttranslational modifications; SMM, stabilized matrix method; SVM, support vector machines; TAP, transporter associated with antigen processing.