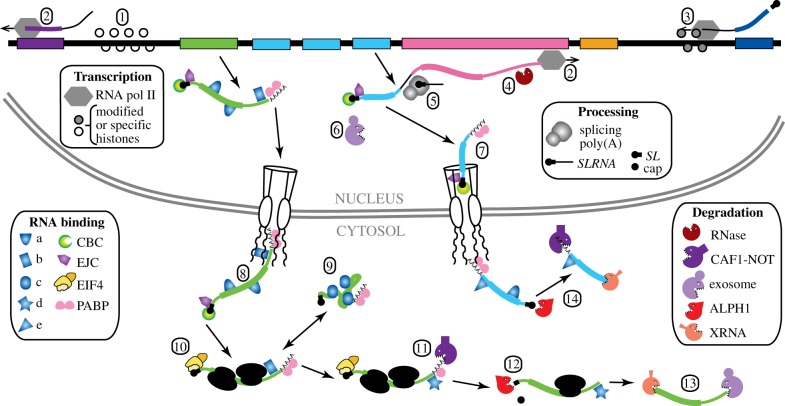
Figure 2.
Gene expression in kinetoplastids. This diagram is modified from [11]. The thick line at the top represents the genome, and each coloured block is represented in a mature mRNA. The corresponding mRNAs are shown with the coding regions thicker than the untranslated regions. The numbered steps are described in the text. 1: Modified histones in an RNA polymerase II initiation region. 2: RNA polymerase II elongation. 3: RNA polymerase II termination. 4: Endonuclease cleavage of precursor (postulated). 5: Trans-splicing and polyadenylation. 6: Incompletely processed mRNAs can be degraded by the exosome. 7: Export of a completed mRNA, with bound poly(A) binding protein (PABP), exon junction complex (EJC) and nuclear cap-binding complex (CBC). (Note that we do not know which, if any, splice junctions are bound by the EJC, and also that mRNA export can commence before the 3′-end is complete.) 8: Emergence of a mature mRNA including proteins on the coding region (a) and a specific, stabilizing protein (b) bound to the 3′-untranslated region (3′-UTR). 9: Binding by a silencing or aggregating RNA-binding protein (c) and condensation into granules. 10: Binding of EIF4E, EIF4G and EIF4A, and translation. 11: Protein (b) is replaced by a destabilizing RNA-binding protein (d) and deadenylation starts. 12: Decapping by ALPH1. 13: Degradation by XRNA and the exosome. 14: Rapid decay pathway—immediate decapping promoted by protein (e). Many of the proteins involved are listed in electronic supplementary material, table S1.