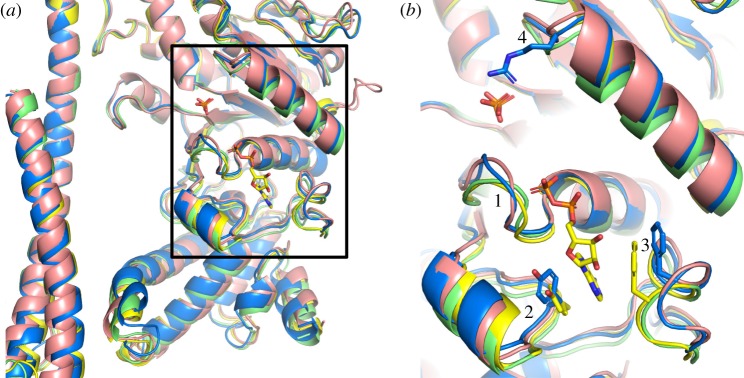
Figure 5.
Comparison of the nucleotide-binding sites in βE-subunits in various bacterial F1-ATPases. (a) Cartoon representation of part of the α-helical coiled-coil of the γ-subunits and adjacent nucleotide-binding domains and C-terminal α-helical domains of βE-subunits based on the superimposition of F1-ATPases via their crown domains; F. nucleatum (6q45; blue); P. denitrificans [54] (5dn6; pink); M. smegmatis [51] (6foc; green); C. thermarum [52] (5ik2; yellow). An ADP molecule bound to the βE-subunit from C. thermarum, and phosphate ions bound to the βE-subunits from C. thermarum and M. smegmatis are shown in the stick representation. (b) Magnified version of the region in the box in (a); regions 1, residues 151–154 towards the N-terminal end of the P-loop (residues 149–156) in F. nucleatum; regions 2 and 3 contain aromatic residues, Tyr-332 and Phe-411 in F. nucleatum (shown in the blue stick representation), that form a pocket where the adenine ring of ADP binds; the equivalent residues in C. thermarum (Tyr-334 and Phe-413) are shown in yellow; region 4, loop containing an arginine residue (Arg-182 from F. nucleatum shown in the blue stick representation) involved in binding phosphate ions in M. smegmatis and C. thermarum, but not evidently in F. nucleatum and P. denitrificans.