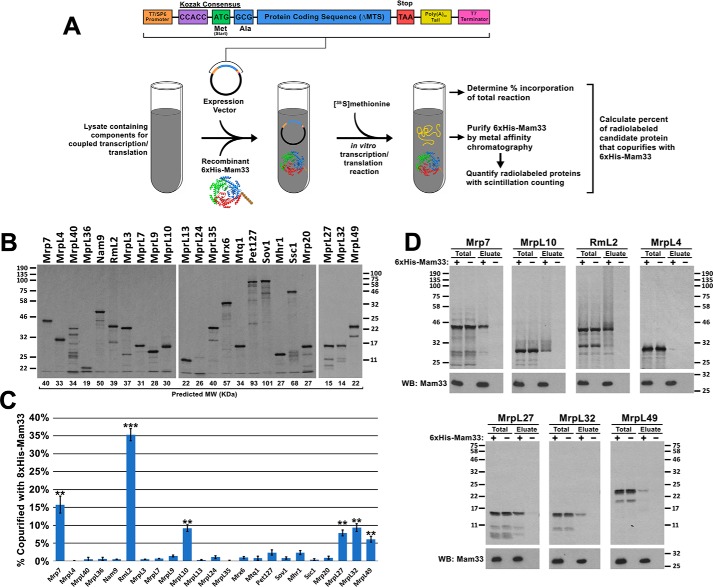
Figure 5.
Mam33 directly interacts with individual mtLSU proteins in vitro. A, workflow: candidate ribosomal proteins from the 8×His-Mam33 affinity purification (Fig. 4) were expressed from a SP6 promoter in either rabbit reticulocyte lysates or wheat germ extracts containing [35S]methionine and recombinant 6×His-Mam33. Following nickel purification, radiolabeled proteins were visualized by autoradiography and quantified by scintillation counting. B, full-length candidate proteins were expressed in the cell-free transcription/translation reaction in the presence of recombinant 6×His-Mam33. Proteins from 1/100 of each reaction were resolved by SDS-PAGE and radiolabeled products were visualized by autoradiography. Predicted molecular weights based on amino acid sequence are shown directly below each lane. Protein molecular masses are indicated in kilodaltons on the left (first two panels) and right (third panel). C, several proteins synthesized in cell-free lysates copurify with recombinant 6×His-Mam33. Percent copurified was calculated as cpm in the bead elution divided by cpm in a TCA precipitate of the total transcription-translation reaction. Bars represent the average of at least three biological replicates minus negative control reactions lacking 6×His-Mam33 performed in parallel. Error bars represent S.E. and two or three asterisks indicate p values smaller than 0.01 or 0.001, respectively (see “Experimental procedures”), for more information on statistical analysis). Raw data are provided in Table S7. D, a fraction of the total reaction and bead eluate from the nickel-agarose beads were visualized by SDS-PAGE and autoradiography for the six interacting proteins (Mrp7, MrpL10, RmL2, MrpL27, MrpL32, MrpL49) and one protein that did not interact (MrpL4). Protein molecular masses are indicated in kilodaltons on the left and right of the upper and lower panel sets.