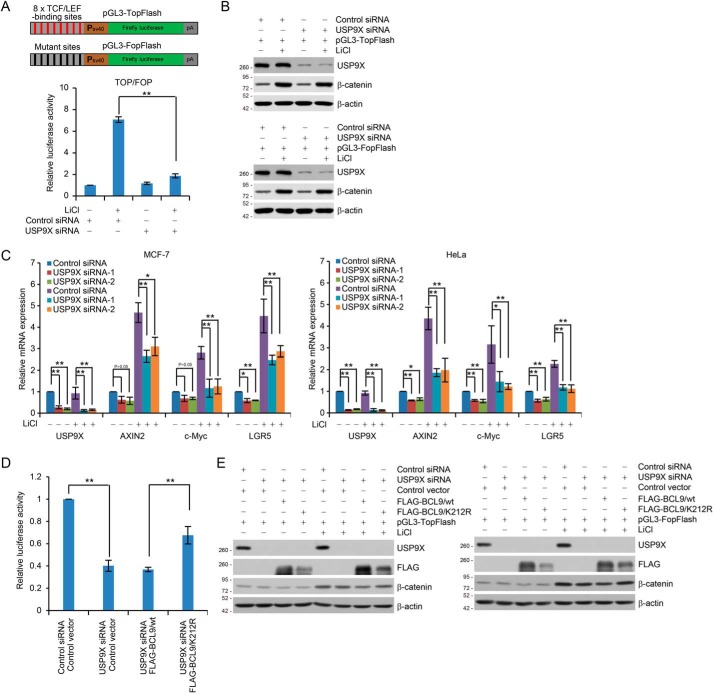
Figure 4.
USP9X-promoted BCL9 deubiquitination is required for transcriptional Wnt responses. A, schematic diagrams of the TopFlash/FopFlash-luciferase reporter constructs are as shown (upper panel). For reporter assays, HEK293T cells were co-transfected with control siRNA or USP9X siRNA together with Renilla and pGL3–TopFlash or pGL3–FopFlash. These cells were cultured in the absence or presence of 25 mm LiCl for 6 h before measuring luciferase activity. The relative luciferase activity was determined by sequential normalization with values of Renilla and pGL3–FopFlash activity. Each bar represents the mean ± S.D. for three biological experiments. **, p < 0.01, one-way ANOVA. B, cellular extracts from cells used in A were analyzed by Western blotting. C, MCF-7 (left panel) or HeLa (right panel) cells transfected with control siRNA or USP9X siRNAs were cultured in the absence or presence of 25 mm LiCl for 6 h and collected for qRT-PCR analysis with USP9X, AXIN2, c-Myc, and LGR5 primers. Each bar represents the mean ± S.D. for three biological experiments. *, p < 0.05; **, p < 0.01, one-way ANOVA. D, HEK293T cells expressing FLAG–BCL9/WT or FLAG–BCL9/K212R were co-transfected with control siRNA or USP9X siRNA together with Renilla and pGL3–TopFlash or pGL3–FopFlash. These cells were cultured in the absence or presence of 25 mm LiCl for 6 h before measuring luciferase activity. The relative luciferase activity was determined by sequential normalization as shown in A and then normalized with the control treatment. Each bar represents the mean ± S.D. for three biological experiments. **, p < 0.01, one-way ANOVA. E, cellular extracts for reporter assays in D were analyzed by Western blotting.