Table 1.
Co-expression analyses for identification of putative UDP-glucose: 12-OH-JA glucosyltransferase genes
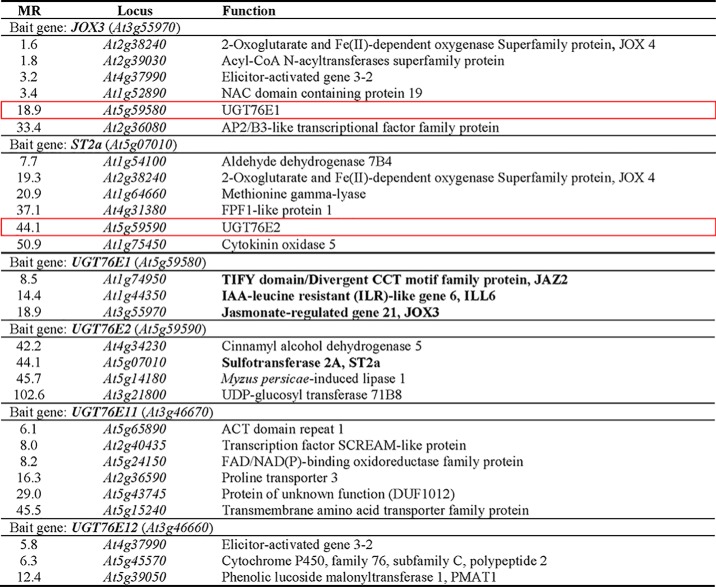
RNA-seq co-expression analysis of genes involved in 12-OH-JA metabolism. Jasmonic acid oxidase 3 (JOX3) and sulfotransferase 2a (ST2a) were used as bait genes to search for UGT genes. Two genes encoding for uncharacterized glycosyltransferases, UGT76E1 and UGT76E2, were identified (red frames). RNA-seq co-expression analysis of genes correlating with UGT76E1, UGT76E2, UGT76E11, and UGT76E12. Identified gene products related to JA metabolism are given in bold. JAZ2, jasmonate-ZIM-domain protein 2; JOX3, jasmonic acid oxidase 3. Given are the complete database outputs for every bait. Gene products are ranked by Mutual Rank (MR), giving an average correlation of two genes indicating stronger correlation by smaller values. Co-expression data are publically available from (32).