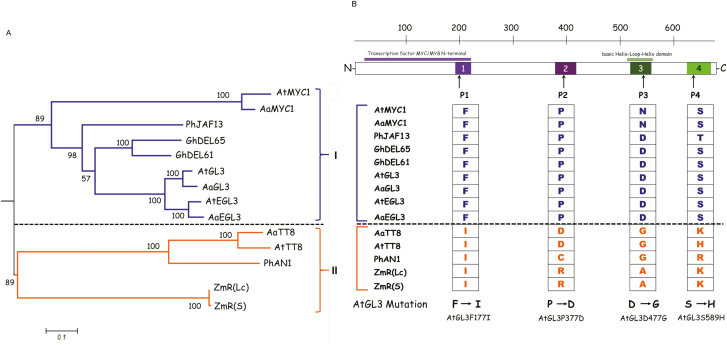
Fig. 4.
Phylogenetic analysis of bHLH proteins used in this study. (A) The phylogenetic tree was constructed using the alignment of full-length bHLH proteins. The tree is drawn to scale with branch lengths measured as the number of substitutions per site, and it was created by mid-point rooting. Bootstrap values are given at the branch nodes, determined from 500 bootstrap repetitions. The interaction behaviors are indicated on the right: clade I, competitive complex formation; clade II, non-competitive complex formation. (B) Analysis of bHLH protein motifs. Four motifs were identified by MEME (http://alternate.meme-suite.org;(Bailey and Gribskov, 1998), shown as numbered boxes in the construct diagram of the bHLH protein. The sequence information for each motif is provided in the Supplementary Fig. S4). The amino acids shared in clade I in the four motifs are shown for the bHLH proteins analysed in this study and compared to those in clade II. The changes in amino acid that were introduced into wild-type AtGL3 are shown at the bottom. (This figure is available in color at JXB online.)